Guide RNA
[2] In bacteria and archaea, gRNAs are a part of the CRISPR-Cas system that serves as an adaptive immune defense that protects the organism from viruses.
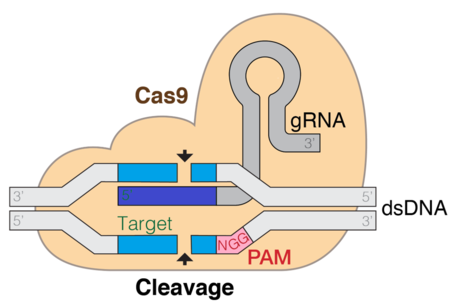
A significant breakthrough occurred in 2012 when it was discovered that gRNA could guide the Cas9 endonuclease to introduce target-specific cuts in double-stranded DNA.
This discovery led to the 2020 Nobel Prize awarded to Jennifer Doudna and Emmanuelle Charpentier for their contributions to the development of CRISPR-Cas9 gene-editing technology.
These frameshifts are corrected post-transcriptionally through the insertion and deletion of uridine residues at precise sites, which then create an open reading frame.
[11] The process of uridine insertion and deletion is mediated by short guide RNAs (gRNAs),which encode the editing information through complementary sequences, and allow for base pairing between guanine and uracil (GU) as well as between guanine and cytosine (GC), facilitating the editing process.
[12] Guide RNAs are mainly transcribed from the intergenic region of DNA maxicircle and have sequences complementary to mRNA.
This pairing recruits a number of ribonucleoprotein complexes that direct the cleavage of the first mismatched base adjacent to the gRNA-mRNA anchor.
The maintenance of editing over the long evolutionary history of these ancient protists suggests the presence of a selective advantage, the exact nature of which is still uncertain.
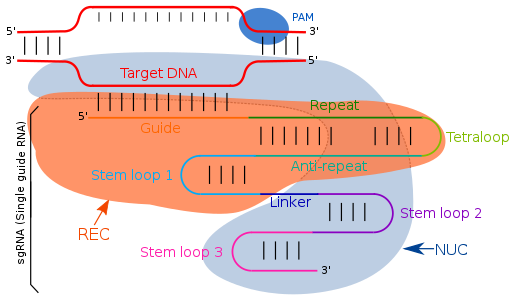
Prokaryotes as bacteria and archaea, use CRISPR (clustered regularly interspaced short palindromic repeats) and its associated Cas enzymes, as their adaptive immune system.
The tracrRNA consist of base pairs that form a stem-loop structure, enabling its attachment to the endonuclease enzyme.
The transcription of the CRISPR locus generates crRNA, which contains spacer regions flanked by repeat sequences, typically 18-20 base pairs (bp) in length.
Modifications in the crRNA sequence within the sgRNA can alter the binding location, allowing for precise targeting of different DNA regions, effectively making it a programmable system for genome editing.
Cas is an endonuclease enzyme that cuts DNA at a specific location directed by a guide RNA.
This is a target-specific technique that can introduce gene knockouts or knock-ins depending on the double strand repair pathway.
The first stage involves the extension of bases in the CRISPR locus region by addition of foreign DNA spacers in the genome sequence.
[32] Guide RNA replaces adenosine with inosine at specific target sites, modifying the genetic code.
[33] Adenosine deaminase acts on RNA, bringing post transcriptional modification by altering codons and different protein functions.