BLAST (biotechnology)
The BLAST program was designed by Eugene Myers, Stephen Altschul, Warren Gish, David J. Lipman and Webb Miller at the NIH and was published in J. Mol.
[5] They proposed "a method for estimating similarities between the known DNA sequence of one organism with that of another",[3] and their work has been described as "the statistical foundation for BLAST.
"[6] Subsequently, Altschul, Gish, Miller, Myers, and Lipman designed and implemented the BLAST program, which was published in the Journal of Molecular Biology in 1990 and has been cited over 100,000 times since.
BLAST is more time-efficient than FASTA by searching only for the more significant patterns in the sequences, yet with comparative sensitivity.
[10] Input sequences (in FASTA or Genbank format), database to search and other optional parameters such as scoring matrix.
If one is attempting to search for a proprietary sequence or simply one that is unavailable in databases available to the general public through sources such as NCBI, there is a BLAST program available for download to any computer, at no cost.
While attempting to find similarity in sequences, sets of common letters, known as words, are very important.
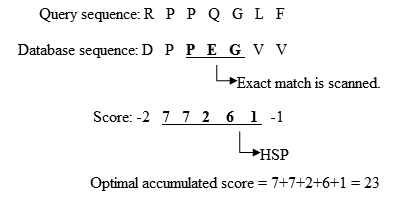
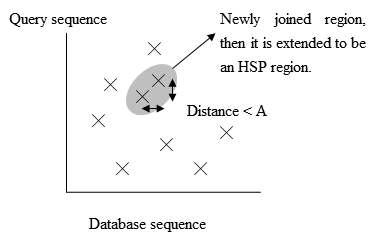
The main idea of BLAST is that there are often High-scoring Segment Pairs (HSP) contained in a statistically significant alignment.
However, the exhaustive Smith-Waterman approach is too slow for searching large genomic databases such as GenBank.
This tool is useful when the reading frame of the DNA sequence is uncertain or contains errors that might cause mistakes in protein-coding.
BLASTx provides combined statistics for hits across all frames, making it helpful for the initial analysis of new DNA sequences.
[18] Parallel BLAST versions of split databases are implemented using MPI and Pthreads, and have been ported to various platforms including Windows, Linux, Solaris, Mac OS X, and AIX.
[20] This allows for significant performance improvements when conducting BLAST searches across a set of nodes in a cluster.
The ideal speed for any parallel computation is a complexity of O(n/p), with n being the size of the database and p being the number of processors.
In addition, the FASTA package provides SSEARCH, a vectorized implementation of the rigorous Smith-Waterman algorithm.
FASTA is slower than BLAST, but provides a much wider range of scoring matrices, making it easier to tailor a search to a specific evolutionary distance.
While BLAST does a linear search, BLAT relies on k-mer indexing the database, and can thus often find seeds faster.
Advances in sequencing technology in the late 2000s has made searching for very similar nucleotide matches an important problem.
Input sequences can then be mapped very quickly, and output is typically in the form of a BAM file.
For protein identification, searching for known domains (for instance from Pfam) by matching with Hidden Markov Models is a popular alternative, such as HMMER.
For applications in metagenomics, where the task is to compare billions of short DNA reads against tens of millions of protein references, DIAMOND[26] runs at up to 20,000 times as fast as BLASTX, while maintaining a high level of sensitivity.
The open-source software MMseqs is an alternative to BLAST/PSI-BLAST, which improves on current search tools over the full range of speed-sensitivity trade-off, achieving sensitivities better than PSI-BLAST at more than 400 times its speed.
However, when compared to BLAST, it is more time consuming and requires large amounts of computing power and memory.
The settings one can change are E-Value, gap costs, filters, word size, and substitution matrix.
These include identifying species, locating domains, establishing phylogeny, DNA mapping, and comparison.