Proteasome
These α subunits are controlled by binding to "cap" structures or regulatory particles that recognize polyubiquitin tags attached to protein substrates and initiate the degradation process.
[3] The proteasomal degradation pathway is essential for many cellular processes, including the cell cycle, the regulation of gene expression, and responses to oxidative stress.
In mammals, the β1, β2, and β5 subunits are catalytic; although they share a common mechanism, they have three distinct substrate specificities considered chymotrypsin-like, trypsin-like, and peptidyl-glutamyl peptide-hydrolyzing (PHGH).
[22] Alternative β forms denoted β1i, β2i, and β5i can be expressed in hematopoietic cells in response to exposure to pro-inflammatory signals such as cytokines, in particular, interferon gamma.
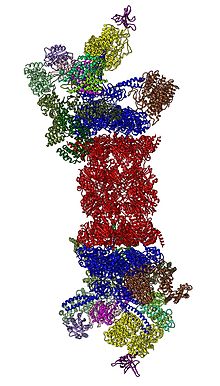
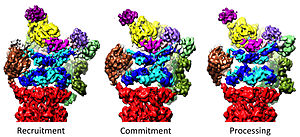
[33][34][35] In 2018, a major effort has elucidated the detailed mechanisms of deubiquitylation, initiation of translocation and processive unfolding of substrates by determining seven atomic structures of substrate-engaged 26S proteasome simultaneously.
Rpn11, the deubiquitinating enzyme, is placed at the mouth of the AAA-ATPase hexamer, ideally positioned to remove ubiquitin moieties immediately before translocation of substrates into the 20S.
[30][31] In the presence of ATP but absence of substrate three alternative, less abundant conformations of the 19S are adopted primarily differing in the positioning of the lid with respect to the AAA-ATPase module.
[42] The expression of the 11S particle is induced by interferon gamma and is responsible, in conjunction with the immunoproteasome β subunits, for the generation of peptides that bind to the major histocompatibility complex.
A few high-resolution snapshots of the proteasome bound to a polyubiquitinated protein suggest that ubiquitin receptors might be coordinated with deubiquitinase Rpn11 for initial substrate targeting and engagement.
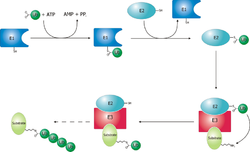
[51] The ubiquitin protein itself is 76 amino acids long and was named due to its ubiquitous nature, as it has a highly conserved sequence and is found in all known eukaryotic organisms.
How Rpn11 removes a ubiquitin chain en bloc from a protein substrate was captured by an atomic structure of the substrate-engaged human proteasome in a conformation named EB.
[57] It appears that USP14 regulates proteasome function at multiple checkpoints by both catalytically competing with Rpn11 and allosterically reprogramming the AAA-ATPase states, which is rather unexpected for a DUB.
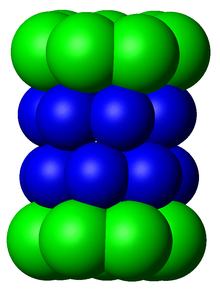
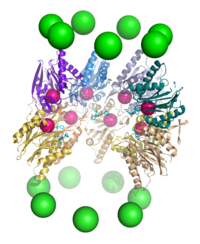
Because the 20S particle's central channel is narrow and gated by the N-terminal tails of the α ring subunits, the substrates must be at least partially unfolded before they enter the core.
[69] Although the three catalytic β subunits have a common mechanism, they have slightly different substrate specificities, which are considered chymotrypsin-like, trypsin-like, and peptidyl-glutamyl peptide-hydrolyzing (PHGH)-like.
Certain transcription factors regulating the expression of specific genes, including one component of the mammalian complex NF-κB, are synthesized as inactive precursors whose ubiquitination and subsequent proteasomal degradation converts them to an active form.
[74] Finally, structurally abnormal, misfolded, or highly oxidized proteins are also subject to ubiquitin-independent and 19S-independent degradation under conditions of cellular stress.
[84] The cellular consequences of ARF activation depend on the plant type and developmental stage, but are involved in directing growth in roots and leaf veins.
The resulting deconstruction of cellular components is primarily carried out by specialized proteases known as caspases, but the proteasome also plays important and diverse roles in the apoptotic process.
The mechanism for this effect is not clear, but is hypothesized to be specific to cells in quiescent states, or to result from the differential activity of the pro-apoptotic kinase JNK.
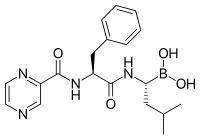
[91] The ability of proteasome inhibitors to induce apoptosis in rapidly dividing cells has been exploited in several recently developed chemotherapy agents such as bortezomib and salinosporamide A.
[95] Oxidized proteins, which often form large amorphous aggregates in the cell, can be degraded directly by the 20S core particle without the 19S regulatory cap and do not require ATP hydrolysis or tagging with ubiquitin.
[98] This hypothesis is supported by the observation that yeast models of Parkinson's are more susceptible to toxicity from α-synuclein, the major protein component of Lewy bodies, under conditions of low proteasome activity.
These proteins whose expression increases during the immune response include the 11S regulatory particle, whose main known biological role is regulating the production of MHC ligands, and specialized β subunits called β1i, β2i, and β5i with altered substrate specificity.
Lactacystin, a natural product synthesized by Streptomyces bacteria, was the first non-peptidic proteasome inhibitor discovered[103] and is widely used as a research tool in biochemistry and cell biology.
[107][108] Preclinical and early clinical studies have been started to examine bortezomib's effectiveness in treating other B-cell-related cancers,[109] particularly some types of non-Hodgkin's lymphoma.
For example, studies in mice bearing human skin grafts found a reduction in the size of lesions from psoriasis after treatment with a proteasome inhibitor.
Accordingly, gene expression by degradation of transcription factors, such as p53, c-jun, c-Fos, NF-κB, c-Myc, HIF-1α, MATα2, STAT3, sterol-regulated element-binding proteins and androgen receptors are all controlled by the UPS and thus involved in the development of various malignancies.
[136] Moreover, the UPS regulates the degradation of tumor suppressor gene products such as adenomatous polyposis coli (APC) in colorectal cancer, retinoblastoma (Rb).
[125] Additionally, the UPS also plays a role in inflammatory responses as regulators of leukocyte proliferation, mainly through proteolysis of cyclines and the degradation of CDK inhibitors.
[137] Lastly, autoimmune disease patients with SLE, Sjögren syndrome and rheumatoid arthritis (RA) predominantly exhibit circulating proteasomes which can be applied as clinical biomarkers.