Residual dipolar coupling
The resulting so-called residual anisotropic magnetic interactions are useful in biomolecular NMR spectroscopy.
As a first example in organic solvents, RDC measurements in stretched polystyrene (PS) gels swollen in CDCl3 were reported as a promising alignment method.
When taken at very high field, these spectra may contain data that can usefully complement NOEs in determining a tertiary fold.
is given by: where The above equation can be rewritten in the following form: where In isotropic solution molecular tumbling reduces the average value of
However the degree of alignment achieved by applying magnetic field is so low that the largest 1H-15N or 1H-13C dipolar couplings are <5 Hz.
However, in practice, this is not achievable and RDC is used mainly to refine a structure determined by NOE data and J-coupling.
Moreover, if a very small set of dipolar couplings are available, the refinement may lead to a structure worse than the original one.
For a protein with N aminoacids, 2N RDC constraint for backbone is the minimum needed for an accurate refinement.
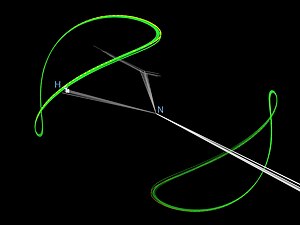
[25] The information content of an individual RDC measurement for a specific bond vector (such as a specific backbone NH bond in a protein molecule) can be understood by showing the target curve that traces out directions of perfect agreement between the observed RDC value and the value calculated from the model.
Such a curve (see figure) has two symmetrical branches that lie on a sphere with its polar axis along the magnetic field direction.
[25] In the case of elongated molecules such as RNA, where local torsional information and short distances are not enough to constrain the structures, RDC measurements can provide information about the orientations of specific chemical bonds throughout a nucleic acid with respect to a single coordinate frame.
Particularly, RNA molecules are proton-poor and overlap of ribose resonances make it very difficult to use J-coupling and NOE data to determine the structure.
RDC measurements have also been proved to be extremely useful for a rapid determination of the relative orientations of units of known structures in proteins.
In particular, due to its radial dependence the RDC is in particular sensitive to large-amplitude angular processes [28] An early example by Tolman et al. found previously published structures of myoglobin insufficient to explain measured RDC data, and devised a simple model of slow dynamics to remedy this.