Salt bridge (protein and supramolecular)
Entropic driving forces for ion pairing (in absence of significant H-bonding contributions) are also found in methanol as solvent.
[1] Although these are the most common, other residues with ionizable side chains such as histidine, tyrosine, and serine can also participate, depending on outside factors perturbing their pKa's.
[12] These contain structures from different enzyme classes, including hydrolase, transferases, kinases, reductase, oxidoreductase, lyases, and G protein-coupled receptors (GPCRs).
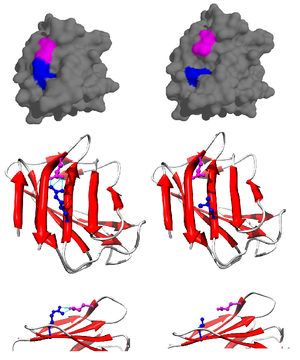
The contribution of a salt bridge to the overall stability to the folded state of a protein can be assessed through thermodynamic data gathered from mutagenesis studies and nuclear magnetic resonance techniques.
[13] Using a mutated pseudo-wild-type protein specifically mutated to prevent precipitation at high pH, the salt bridge’s contribution to the overall free energy of the folded protein state can be determined by performing a point-mutation, altering and, consequently, breaking the salt bridge.
This is quantified through a method described by Becktel and Schellman where the free energy difference between the two is calculated through ΔTΔS.
The second method utilizes nuclear magnetic resonance spectroscopy to calculate the free energy of the salt bridge.
A titration is performed, while recording the chemical shift corresponding to the protons of the carbon adjacent to the carboxylate or ammonium group.
In the unfolded wild-type protein, where the salt bridge is absent, His31 is reported to have a pKa of 6.8 in H2O buffers of moderate ionic strength.
When the salt bridge is disrupted, like in the mutant D70N, the pKa shifts back to a value of 6.9, much closer to that of His31 in the unfolded state.
Entropy plays a larger role in surface salt bridges where residues that normally have the ability to move are constricted by their electrostatic interaction and hydrogen bonding.
Salt bridges have been used by chemists within this field in both diverse and creative ways, including sensing of anions, the synthesis of molecular capsules and double helical polymers.
[18][19][20][21][22][23] Ion pairing is the most important driving force for anion complexation, but selectivity e.g. within the halide series has been achieved, mostly by hydrogen bonds contributions.
Yashima and coworkers have used salt bridges to construct several polymers that adopt a double helix conformation much like DNA.
[26] Starting from their monomer and platinum(II) biphenyl (Figure 8), their metallopolymer self assembles through a series of ligand exchange reactions.