Three prime untranslated region
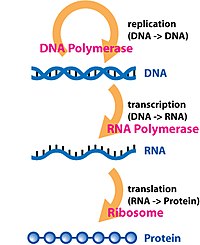
In molecular genetics, the three prime untranslated region (3′-UTR) is the section of messenger RNA (mRNA) that immediately follows the translation termination codon.
By binding to specific sites within the 3′-UTR, miRNAs can decrease gene expression of various mRNAs by either inhibiting translation or directly causing degradation of the transcript.
Furthermore, the 3′-UTR contains the sequence AAUAAA that directs addition of several hundred adenine residues called the poly(A) tail to the end of the mRNA transcript.
For example, poly(A) tail bound PABP interacts with proteins associated with the 5' end of the transcript, causing a circularization of the mRNA that promotes translation.
The 3′-UTR can also contain sequences that attract proteins to associate the mRNA with the cytoskeleton, transport it to or from the cell nucleus, or perform other types of localization.
In addition to sequences within the 3′-UTR, the physical characteristics of the region, including its length and secondary structure, contribute to translation regulation.
One possible explanation for this phenomenon is that longer regions have a higher probability of possessing more miRNA binding sites that have the ability to inhibit translation.
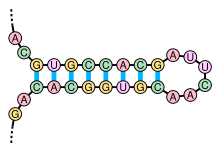
The IRE is a stem-loop structure within the untranslated regions of mRNAs that encode proteins involved in cellular iron metabolism.
The mRNA transcript containing this element is either degraded or stabilized depending upon the binding of specific proteins and the intracellular iron concentrations.
[4] The 3′-untranslated region plays a crucial role in gene expression by influencing the localization, stability, export, and translation efficiency of an mRNA.
miRNAs are short, non-coding RNA molecules capable of binding to mRNA transcripts and regulating their expression.
[1][2] While the presence of a poly(A) tail usually aids in triggering translation, the absence or removal of one often leads to exonuclease-mediated degradation of the mRNA.
In general, longer 3′-UTRs correspond to lower expression rates since they often contain more miRNA and protein binding sites that are involved in inhibiting translation.
The most common structure is a stem-loop, which provides a scaffold for RNA binding proteins and non-coding RNAs that influence expression of the transcript.
Since it can affect the presence of protein and miRNA binding sites, APA can cause differential expression of mRNA transcripts by influencing their stability, export to the cytoplasm, and translation efficiency.
[9] Dysregulation of ARE-binding proteins (AUBPs) due to mutations in AU-rich regions can lead to diseases including tumorigenesis (cancer), hematopoietic malignancies, leukemogenesis, and developmental delay/autism spectrum disorders.
[10][11][12] An expanded number of trinucleotide (CTG) repeats in the 3’-UTR of the dystrophia myotonica protein kinase (DMPK) gene causes myotonic dystrophy.
[7] Elements in the 3′-UTR have also been linked to human acute myeloid leukemia, alpha-thalassemia, neuroblastoma, Keratinopathy, Aniridia, IPEX syndrome, and congenital heart defects.
[7] Future research through the increased use of deep-sequencing based ribosome profiling will reveal more regulatory subtleties as well as new control elements and AUBPs.