BioBrick
[1][2] These building blocks are used to design and assemble larger synthetic biological circuits from individual parts and combinations of parts with defined functions, which would then be incorporated into living cells such as Escherichia coli cells to construct new biological systems.
[3] Examples of BioBrick parts include promoters, ribosomal binding sites (RBS), coding sequences and terminators.
There are three levels to the hierarchy: The development of standardized biological parts allows for the rapid assembly of sequences.
[1] Since then, various research groups have utilized the BioBrick standard parts to engineer novel biological devices and systems.
The BioBricks Foundation was formed in 2006 by engineers and scientists alike as a not-for-profit organization to standardize biological parts across the field.
[6] The Foundation focuses on improving in areas of Technology, Law, Education and the Global Community as they apply to synthetic biology.
BioBricks Foundation's activities include hosting SBx.0 Conferences, technical and educational programs.
Those who want to give a part to the community sign the Contributor Agreement, agreeing not to assert against Users Contributor-held intellectual property rights that might limit the use of the contributed materials.
The BioBrick assembly standard is a more reliable approach for combining parts to form larger composites.
Besides that, when compared to the old-fashioned ad hoc cloning method, the assembly standard process is faster and promotes automation.
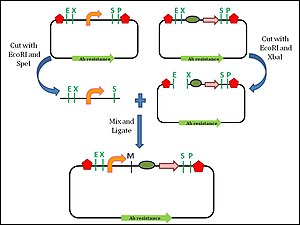
Every BioBrick part is a DNA sequence which is carried by a circular plasmid, which acts as a vector.
[13] The prefix and suffix sequences remain unchanged by this digestion and ligation process, which allows for subsequent assembly steps with more BioBrick parts.
This assembly is an idempotent process: multiple applications do not change the end product, and maintain the prefix and suffix.
[12] The scar site causes a frame shift which prevents the continuous reading of codons, which is required for the formation of fusion protein.
Tom Knight later developed the BB-2 assembly standard in 2008 to address problems with joining the scars of protein domains and that the scars consist of eight bases, which will yield an altered reading frame when joining protein domains.
[14] The BglBrick assembly standard was proposed by J. Christopher Anderson, John E. Dueber, Mariana Leguia, Gabriel C. Wu, Jonathan C. Goler, Adam P. Arkin, and Jay D. Keasling in September 2009 as a standard very similar in concept to BioBrick, but enabling the generation of fusion proteins without altering the reading frame or introducing stop codons and while creating a relatively neutral amino acid linker scar (GlySer).
The assembly technique proposed by the Freiburg team diminishes the limitations of the Biofusion standard.
It also has a higher transformation rate than 3A assembly, and it does not require the involved plasmids to have different antibiotic resistance genes.
The plasmids are reacted in vivo with sequence-specific DNA methyltransferases, so that each is modified and protected from one of two restriction endonucleases that are later used to linearize undesired circular ligation products.
BIOFAB aims to catalogue high-quality BioBrick parts to accommodate the needs of professional synthetic biology community.
The BBF is currently working on the derivation of standard framework to promote the production high quality BioBrick parts which would be freely available to everyone.