Cas9
The CRISPR-Cas9 genome editing technique was a significant contributor to the Nobel Prize in Chemistry in 2020 being awarded to Emmanuelle Charpentier and Jennifer Doudna.
[2] More technically, Cas9 is a RNA-guided DNA endonuclease enzyme associated with the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) adaptive immune system in Streptococcus pyogenes.
[4][6][7][8] Cas9 performs this interrogation by unwinding foreign DNA and checking for sites complementary to the 20 nucleotide spacer region of the guide RNA (gRNA).
Apart from its original function in bacterial immunity, the Cas9 protein has been heavily utilized as a genome engineering tool to induce site-directed double-strand breaks in DNA.
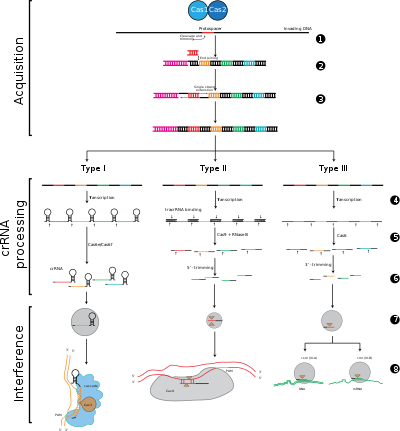
[14] To survive in a variety of challenging, inhospitable habitats that are filled with bacteriophages, bacteria and archaea have evolved methods to evade and fend off predatory viruses.
In 2005, it was discovered by three separate groups that the spacer regions were homologous to foreign DNA elements, including plasmids and viruses.
[20] Interference involves the crRNAs within a multi-protein complex called CASCADE, which can recognize and specifically base-pair with regions of inserting complementary foreign DNA.
The crRNA-foreign nucleic acid complex is then cleaved, however if there are mismatches between the spacer and the target DNA, or if there are mutations in the PAM, then cleavage will not be initiated.
In the latter scenario, the foreign DNA is not targeted for attack by the cell, thus the replication of the virus proceeds and the host is not immune to viral infection.
Protospacers and protospacer-associated motifs (shown in red) are acquired at the "leader" end of a CRISPR array in the host DNA.
The CRISPR array is composed of spacer sequences (shown in colored boxes) flanked by repeats (black diamonds).
crRNA with a spacer that has strong complementarity to the incoming foreign DNA begins a cleavage event (depicted with scissors), which requires Cas proteins.
The interference stage can be functionally and temporarily distinct from CRISPR acquisition and expression (depicted by white line dividing the cell).
Since dCas9 appears to down regulate gene expression, this action is amplified even more when it is used in conjunction with repressive chromatin modifier domains.
A promoter can be added to the dCas9 protein which allows them to work with each other to become efficient at beginning or stopping transcription at different sequences along a strand of DNA.
This is prevalent in certain types of prokaryotes when a promoter and dCas9 align themselves together to impede the ability of elongation of polymer of nucleotides coming together to form a transcribed piece of DNA.
[23] When examining the effects of repression of transcription further, H3K27, an amino acid component of a histone, becomes methylated through the interaction of dCas9 and a peptide called FOG1.
When the dCas9 attaches to a form of RNA called guide-RNA, it prevents the proliferation of repeating codons and DNA sequences that might be harmful to an organism's genome.
A key feature of the target DNA is that it must contain a protospacer adjacent motif (PAM) consisting of the three-nucleotide sequence- NGG.
REC1 and BH related mutants show lower or none activity compared with wild type, which indicate these two domains are crucial for the sgRNA recognition at repeat sequence and stabilization of the whole complex.
Although the interactions between spacer sequence and Cas9 as well as PI domain and repeat region need further studies, the co-crystal demonstrates clear interface between Cas9 and sgRNA.
[citation needed] Wild-type S. pyogenes Cas9 requires magnesium (Mg2+) cofactors for the RNA-mediated DNA cleavage; however, Cas9 has been shown to exhibit varying levels of activity in the presence of other divalent metal ions.
[32] The kinetics of DNA cleavage by Cas9 have been of great interest to the scientific community, as this data provides insight into the intricacies of the reaction.
[36][37] The sequence composition at the target DNA site complementary to the 20 nucleotide spacer region of the gRNA also affects cleavage efficiency.
[43] Using molecular dynamics simulation, a study reported that cleavage of the NTS between 17|16 of the target sequence was more energetically favored than 18|17, generating 1 nucleotide 5’ ssDNA overhangs.
The association between staggered cleavage and precise templated insertions was supported by additional studies in human cells.
[45][46] Recently, a high-throughput investigation of Cas9 scission profile revealed that ~85% of on-target cleavage is blunt, whereas ~15% had a 1 nucleotide 5' overhang.
To accomplish this, the two crucial catalytic residues of the RuvC and HNH domain can be mutated to alanine abolishing all endonuclease activity of Cas9.
Beyond direct binding of dCas9 to transcriptionally sensitive positions of loci, dCas9 can be fused to a variety of modulatory protein domains to carry out a myriad of functions.
By employing large libraries of guide RNAs capable of targeting thousands of genes, genome wide genetic screens using dCas9 have been conducted.