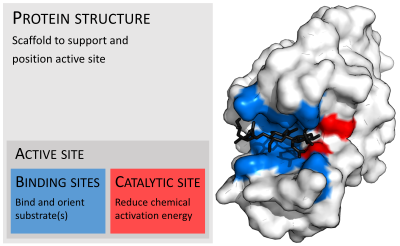
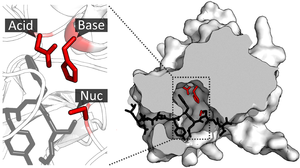
Active site
In biology and biochemistry, the active site is the region of an enzyme where substrate molecules bind and undergo a chemical reaction.
Although the active site occupies only ~10–20% of the volume of an enzyme,[1]: 19 it is the most important part as it directly catalyzes the chemical reaction.
[2] Each active site is evolved to be optimised to bind a particular substrate and catalyse a particular reaction, resulting in high specificity.
To maintain this defined three-dimensional structure, proteins rely on various types of interactions between their amino acid residues.
[citation needed] A tighter fit between an active site and the substrate molecule is believed to increase the efficiency of a reaction.
If one substrate perfectly binds to its active site, the interactions between them will be strongest, resulting in high catalytic efficiency.
The Lock and Key hypothesis cannot explain this, as it would predict a high efficiency of methylglucoside glycosyl transfer due to its tight binding.
For example, when a carboxylic acid (R-COOH) dissociates into RCOO− and H+ ions, COO− will attract positively charged groups such as protonated guanidine side chain of arginine.
Although the individual force is weak, as the total number of interactions between the active site and substrate is massive the sum of them will be significant.
[citation needed] Catalytic residues of the site interact with the substrate to lower the activation energy of a reaction and thereby make it proceed faster.
[citation needed] During enzyme catalytic reaction, the substrate and active site are brought together in a close proximity.
In addition, this binding is favoured by entropy as the energy cost associated with solution reaction is largely eliminated since solvent cannot enter active site.
But if the transition state involves the formation of an ion centre then the side chain will now produce a favourable interaction.
These acids and bases can stabilise the nucleophile or electrophile formed during the catalysis by providing positive and negative charges.
The formation of transition state within the solution requires a large amount of energy to relocate solvent molecules and the reaction is slowed.
So the active site can substitute solvent molecules and surround the substrates to minimize the counterproductive effect imposed by the solution.
The presence of charged groups with the active site will attract substrates and ensure electrostatic complementarity.
During this process, its thiol side chain is oxidised and two glutathione molecules are connected by a disulphide bond to form a dimer(GSSG).
In the active site, there are two cysteine residues besides the FAD cofactor and are used to break the disulphide bond during the catalytic reaction.
This S− group will act as a nucleophile to attack the disulphide bond in the oxidised glutathione(GSSG), breaking it and forming a cysteine-SG complex.
The resulting hydroxide anion nucleophilically attacks the acyl-enzyme complex to form a second tetrahedral oxyanion intermediate, which is once again stabilised by H bonds.
Larger ligands generally stay in the active site longer,[12] as do those with more rotatable bonds (although this may be a side effect of size).
As a result, they can fit into the active site and trigger favourable interactions to fill in the space and block substrates from entry.
HIV protease is used by the virus to cleave Gag-Pol polyprotein into 3 smaller proteins that are responsible for virion assembly, package and maturation.
Since they share a similar structure and electrostatic arrangement to the transition state of substrates they can still fit into the active site but cannot be broken down, so hydrolysis cannot occur.
Glycine can inhibit the activity of neurotransmitter receptors, thus a larger amount of acetylcholinesterase is required to trigger an action potential.
It inhibits glycine receptors(a chloride channel) and a much lower level of neurotransmitter concentration can trigger an action potential.
The OH group in the active site acts as a nucleophile to attack the phosphorus in DIFP and form a tetrahedral intermediate and release a proton.
A covalent bond formed between the active site and DIFP, so the serine side chain is no longer available to the substrate.
This involves the description of the size of an active site and the number and properties of sub-sites, such as details of the binding interaction.