Enhancer RNA
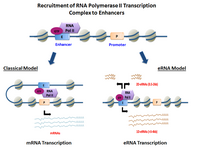
[4] Increasing evidence suggests that eRNAs actively play a role in transcriptional regulation in cis and in trans, and while their mechanisms of action remain unclear, a few models have been proposed.
[6] In parallel studies, 4,588 high confidence extragenic RNA Pol II binding sites were identified in murine macrophages stimulated with the inflammatory mediater lipopolysaccharide to induce transcription.
[2] These eRNAs, unlike messenger RNAs (mRNAs), lacked modification by polyadenylation, were generally short and non-coding, and were bidirectionally transcribed.
The nature of the pre-initiation complex and specific transcription factors recruited to the enhancer may control the type of eRNAs generated.
These long non-coding RNAs, which accurately reflect the host gene's structure except for the alternative first exon, display poor coding potential.
Carullo et al.[13] used these cultured neurons to examine the timing of specific enhancer eRNAs compared to the mRNAs of their target genes.
[14] The functions for eRNA described below have been reported in diverse biological systems, often demonstrated with a small number of specific enhancer-target gene pairs.
P-TEFb can also phosphorylate the negative elongation factor NELF (which pauses RNAP II within 60 nucleotides after mRNA initiation begins).
Since multiple studies have shown that RNA Pol II can be found at a very large number of extragenic regions, it is possible that eRNAs simply represent the product of random “leaky” transcription and carry no functional significance.
It was proposed that the presence of these enzymes could also induce an opening of chromatin at enhancer regions, which are usually present at distant locations but can be recruited to target genes through looping of DNA.
Supporting this hypothesis, transcripts originating from enhancers upstream of the Cyclin D1 gene are thought to serve as adaptors for the recruitment of histone acetyltransferases.
The detection of eRNAs is fairly recent (2010) and has been made possible through the use of genome-wide investigation techniques such as RNA sequencing (RNA-seq) and chromatin immunoprecipitation-sequencing (ChIP-seq).
[1] RNA-seq permits the direct identification of eRNAs by matching the detected transcript to the corresponding enhancer sequence through bioinformatic analyses.
[27] Although some data remain controversial, the consensus in the literature is that the best combination of histone post-translational modifications at active enhancers is made of H2AZ, H3K27ac, and a high ratio of H3K4me1 over H3K4me3.
[5] The experimental detection of eRNAs is complicated by their low endogenous stability conferred by exosome degradation and nonsense-mediated decay.
Evidence that eRNAs cause downstream effects on the efficiency of enhancer activation and gene transcription suggests its functional capabilities and potential importance.
[33] These p53-bound enhancer regions (p53BERs) are shown to interact with multiple local and distal gene targets involved in cell proliferation and survival.
With the emergence of eRNAs as important components in enhancer activity, powerful therapeutic tools such as RNAi may provide promising routes to target disruption of gene expression.