Enhancer (genetics)
In genetics, an enhancer is a short (50–1500 bp) region of DNA that can be bound by proteins (activators) to increase the likelihood that transcription of a particular gene will occur.
[11] Since 2022, scientists have used artificial intelligence to design synthetic enhancers and applied them in animal systems, first in a cell line,[12] and one year later also in vivo.
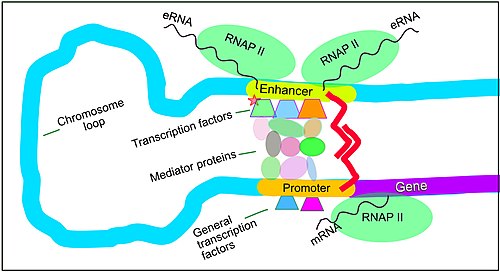
[16] Enhancers do not act on the promoter region itself, but are bound by activator proteins as first shown by in vivo competition experiments.
[33] The schematic illustration in this section shows an enhancer looping around to come into close physical proximity with the promoter of a target gene.
[43] The GADD45G regulator in mice and chimps is active in regions of the brain where cells that form the cortex, ventral forebrain, and thalamus are located and may suppress further neurogenesis.
[citation needed] The development, differentiation and growth of cells and tissues require precisely regulated patterns of gene expression.
In genetically tractable models such as the fruit fly Drosophila melanogaster, for example, a reporter construct such as the lacZ gene can be randomly integrated into the genome using a P element transposon.
Next-generation sequencing (NGS) methods now enable high-throughput functional CRM discovery assays, and the vastly increasing amounts of available data, including large-scale libraries of transcription factor-binding site (TFBS) motifs, collections of annotated, validated CRMs, and extensive epigenetic data across many cell types, are making accurate computational CRM discovery an attainable goal.
An example of NGS-based approach called DNase-seq have enabled identification of nucleosome-depleted, or open chromatin regions, which can contain CRM.
Nucelosome depleted regions can be identified in vivo through expression of Dam methylase, allowing for greater control of cell-type specific enhancer identification.
[46] Computational methods include comparative genomics, clustering of known or predicted TF-binding sites, and supervised machine-learning approaches trained on known CRMs.
All of these methods have proven effective for CRM discovery, but each has its own considerations and limitations, and each is subject to a greater or lesser number of false-positive identifications.
mRNA expression of the reporter can be visualized by in situ hybridization, which provides a more direct measure of enhancer activity, since it is not subjected to the complexities of translation and protein folding.
Thus, unique combinations of pair-rule gene expression create spatial domains along the anterior-posterior axis to set up each of the 14 individual segments.
The 480 bp enhancer responsible for driving the sharp stripe two of the pair-rule gene even-skipped (eve) has been well-characterized.
Gata4 expression is controlled in the early embryo by an intronic enhancer that binds another forkhead domain transcription factor, FoxA2.
Recent work has shown that multiple enhancers allow fruit flies to survive environmental perturbations, such as an increase in temperature.
[citation needed] Recent work has investigated the role of enhancers in morphological changes in threespine stickleback fish.
A more thorough characterization showed that a 500 base pair enhancer sequence is responsible for turning on Pitx1 expression in the posterior fin bud.
[57] Each cell typically contains several hundred of a special class of enhancers that stretch over many kilobases long DNA sequences, called "super-enhancers".
[58] These enhancers contain a large number of binding sites for sequence-specific, inducible transcription factors, and regulate expression of genes involved in cell differentiation.
[62][63] In cancer, proteins that control NF-κB activity are dysregulated, permitting malignant cells to decrease their dependence on interactions with local tissue, and hindering their surveillance by the immune system.
Since 2022, artificial intelligence and transfer learning strategies have led to a better understanding of the features of regulatory DNA sequences, the prediction, and the design of synthetic enhancers.
[66][67] Building on work in cell culture,[66] synthetic enhancers were successfully applied to entire living organisms in 2023.
Using deep neural networks, scientists simulated the evolution of DNA sequences to analyze the emergence of features that underlie enhancer function.
This allowed the design and production of a range of functioning synthetic enhancers for different cell types of the fruit fly brain.
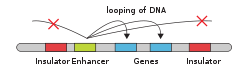

- DNA
- Enhancer
- Promoter
- Gene
- Transcription Activator Protein
- Mediator Protein
- RNA Polymerase