Interactome
The word "interactome" was originally coined in 1999 by a group of French scientists headed by Bernard Jacq.
Molecular interactions can occur between molecules belonging to different biochemical families (proteins, nucleic acids, lipids, carbohydrates, etc.)
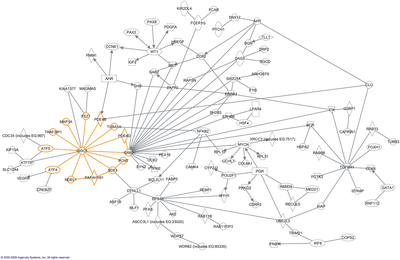
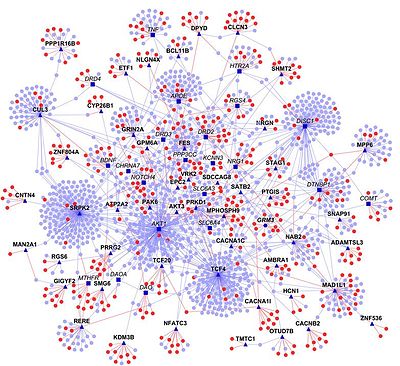
Most commonly, interactome refers to protein–protein interaction (PPI) network (PIN) or subsets thereof.
From this model genetic interactions can be observed at multiple scales which will assist in the study of concepts such as gene conservation.
Interactomics is an example of "top-down" systems biology, which takes an overhead view of a biosystem or organism.
Large sets of genome-wide and proteomic data are collected, and correlations between different molecules are inferred.
While there are numerous methods to study PPIs, there are relatively few that have been used on a large scale to map whole interactomes.
Yeast two hybrid screens allow false positive interactions between proteins that are never expressed in the same time and place; affinity capture mass spectrometry does not have this drawback, and is the current gold standard.
Such analyses are mainly carried out using bioinformatics methods and include the following, among many others: First, the coverage and quality of an interactome has to be evaluated.
[13] The coverage of an interactome can be assessed by comparing it to benchmarks of well-known interactions that have been found and validated by independent assays.
[15] Using experimental data as a starting point, homology transfer is one way to predict interactomes.
[21][22] Other algorithms use only sequence information, thereby creating unbiased complete networks of interaction with many mistakes.
[16][24] Random Forest has been found to be most-effective machine learning method for protein interaction prediction.
[16] Some efforts have been made to extract systematically interaction networks directly from the scientific literature.
Such approaches range in terms of complexity from simple co-occurrence statistics of entities that are mentioned together in the same context (e.g. sentence) to sophisticated natural language processing and machine learning methods for detecting interaction relationships.
[30] The topology of an interactome makes certain predictions how a network reacts to the perturbation (e.g. removal) of nodes (proteins) or edges (interactions).
[31] Such perturbations can be caused by mutations of genes, and thus their proteins, and a network reaction can manifest as a disease.
Network properties include the degree distribution, clustering coefficients, betweenness centrality, and many others.
In contrast, "date hubs" do not exhibit such a correlation and appear to connect different functional modules.
The resultant decline in the efficiency of selection seems to be sufficient to influence a wide range of attributes at the genomic level in a nonadaptive manner.
The Nature study shows that the variation in the power of random genetic drift is also capable of influencing phylogenetic diversity at the subcellular and cellular levels.
Thus, population size would have to be considered as a potential determinant of the mechanistic pathways underlying long-term phenotypic evolution.
In the study it is further shown that a phylogenetically broad inverse relation exists between the power of drift and the structural integrity of protein subunits.
By this means, the complex protein architectures and interactions essential to the genesis of phenotypic diversity may initially emerge by non-adaptive mechanisms.
However, some authors have argued that such non-reproducibility results from the extraordinary sensitivity of various methods to small experimental variation.
While genomes are stable, interactomes may vary between tissues, cell types, and developmental stages.
Each protein–protein interactome may represent only a partial sample of potential interactions, even when a supposedly definitive version is published in a scientific journal.
The binding strength of the various protein interactors, microenvironmental factors, sensitivity to various procedures, and the physiological state of the cell all impact protein–protein interactions, yet are usually not accounted for in interactome studies.