List of biophysically important macromolecular crystal structures
Crystal structures of protein and nucleic acid molecules and their complexes are central to the practice of most parts of biophysics, and have shaped much of what we understand scientifically at the atomic-detail level of biology.
Their importance is underlined by the United Nations declaring 2014 as the International Year of Crystallography, as the 100th anniversary of Max von Laue's 1914 Nobel Prize for discovering the diffraction of X-rays by crystals.
[1] The list includes all the first dozen distinct structures, those that broke new ground in subject or method, and those that became model systems for work in future biophysical areas of research.
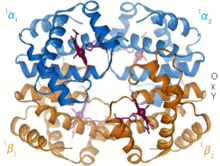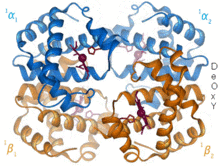
1960 – The hemoglobin crystal structure[4] showed a tetramer of two related chain types and was solved at much lower resolution than the monomeric myoglobin, but it clearly had the same basic 8-helix architecture (now called the "globin fold").
[10] The starting material for preparation can be bought at the grocery store, and hen-egg lysozyme crystallizes very readily in many different space groups; it is the favorite test case for new crystallographic experiments and instruments.
Later a small protein inhibitor of carboxypeptidase was solved (PDB file 4CPA)[22] that mechanically stops the catalysis by presenting its C-terminal end just sticking out from between a ring of disulfide bonds with tight structure behind it, preventing the enzyme from sucking in the chain past the first residue.
1970 – Rubredoxin (PDB file 2rxn[26]) was the first redox structure solved, a minimalist protein with the iron bound by 4 Cys sidechains from 2 loops at the top of β hairpins.
Insulin was a success of NASA's crystal-growth program on the Space Shuttle, producing bulk preparations of very uniform tiny crystals for controlled dosage.