Molecular models of DNA
From the very early stages of structural studies of DNA by X-ray diffraction and biochemical means, molecular models such as the Watson-Crick nucleic acid double helix model were successfully employed to solve the 'puzzle' of DNA structure, and also find how the latter relates to its key functions in living cells.
The first high quality X-ray diffraction patterns of A-DNA were reported by Rosalind Franklin and Raymond Gosling in 1953.
[1] Rosalind Franklin made the critical observation that DNA exists in two distinct forms, A and B, and produced the sharpest pictures of both through X-ray diffraction technique.
The first reports of a double helix molecular model of B-DNA structure were made by James Watson and Francis Crick in 1953.
Wilson, reported the first X-ray patterns of in vivo B-DNA in partially oriented salmon sperm heads.
[7] The development of the first correct double helix molecular model of DNA by Crick and Watson may not have been possible without the biochemical evidence for the nucleotide base-pairing ([A---T]; [C---G]), or Chargaff's rules.
Epigenetic transformation studies of DNA in vivo were however much slower to develop despite their importance for embryology, morphogenesis and cancer research.
Although DNA consists of relatively rigid, very large elongated biopolymer molecules called fibers or chains (that are made of repeating nucleotide units of four basic types, attached to deoxyribose and phosphate groups), its molecular structure in vivo undergoes dynamic configuration changes that involve dynamically attached water molecules and ions.
Such varying molecular geometries can also be computed, at least in principle, by employing ab initio quantum chemistry methods that can attain high accuracy for small molecules, although claims that acceptable accuracy can be also achieved for polynuclelotides, and DNA conformations, were recently made on the basis of vibrational circular dichroism (VCD) spectral data.
Such quantum geometries define an important class of ab initio molecular models of DNA which exploration has barely started, especially related to results obtained by VCD in solutions.
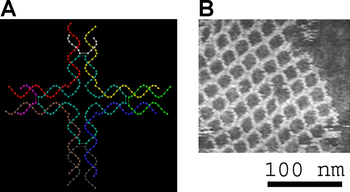
Animated molecular models, such as the wire, or skeletal, type shown at the top of this article, allow one to visually explore the three-dimensional (3D) structure of DNA.
The hydrogen bonding dynamics and proton exchange is very different by many orders of magnitude between the two systems of fully hydrated DNA and water molecules in ice.
Experimental methods which can directly measure the mechanical properties of DNA are relatively new, and high-resolution visualization in solution is often difficult.
[14][15] After DNA has been separated and purified by standard biochemical methods, one has a sample in a jar much like in the figure at the top of this article.