Mutation
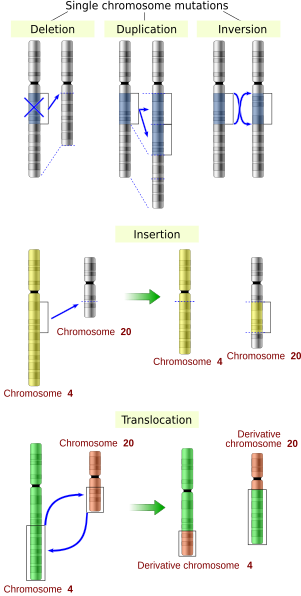
Mutations may also result from substitution, insertion or deletion of segments of DNA due to mobile genetic elements.
Mutation is the ultimate source of all genetic variation, providing the raw material on which evolutionary forces such as natural selection can act.
A 2007 study on genetic variations between different species of Drosophila suggested that, if a mutation changes a protein produced by a gene, the result is likely to be harmful, with an estimated 70% of amino acid polymorphisms that have damaging effects, and the remainder being either neutral or marginally beneficial.
[24] Another effect of these mobile DNA sequences is that when they move within a genome, they can mutate or delete existing genes and thereby produce genetic diversity.
[38] Likewise, in yeast, Kunz et al.[39] found that more than 60% of the spontaneous single base pair substitutions and deletions were caused by translesion synthesis.
As S. Rosenberg states, "These mechanisms reveal a picture of highly regulated mutagenesis, up-regulated temporally by stress responses and activated when cells/organisms are maladapted to their environments—when stressed—potentially accelerating adaptation.
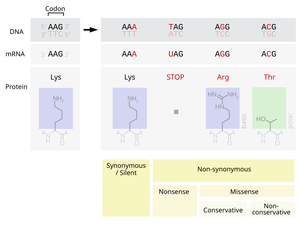
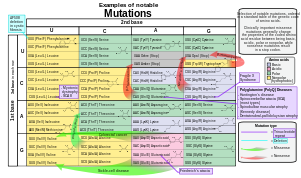
[47] Gene mutations have varying effects on health depending on where they occur and whether they alter the function of essential proteins.
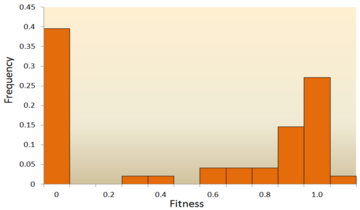
For instance, in a screen of all gene deletions in E. coli, 80% of mutations were negative, but 20% were positive, even though many had a very small effect on growth (depending on condition).
Attempts have been made to infer the distribution of fitness effects (DFE) using mutagenesis experiments and theoretical models applied to molecular sequence data.
DFE, as used to determine the relative abundance of different types of mutations (i.e., strongly deleterious, nearly neutral or advantageous), is relevant to many evolutionary questions, such as the maintenance of genetic variation,[62] the rate of genomic decay,[63] the maintenance of outcrossing sexual reproduction as opposed to inbreeding[64] and the evolution of sex and genetic recombination.
[93] In order to categorize a mutation as such, the "normal" sequence must be obtained from the DNA of a "normal" or "healthy" organism (as opposed to a "mutant" or "sick" one), it should be identified and reported; ideally, it should be made publicly available for a straightforward nucleotide-by-nucleotide comparison, and agreed upon by the scientific community or by a group of expert geneticists and biologists, who have the responsibility of establishing the standard or so-called "consensus" sequence.
In sexually reproducing organisms, the comparatively higher frequency of cell divisions in the parental sperm donor germline drive conclusions that rates of de novo mutation can be tracked along a common basis.
[105] This claim combines the observed effects of increased probability for mutation in rapid spermatogenesis with short periods of time between cellular divisions that limit the efficiency of repair machinery.
For example, certain intensities of exposure to radioactive elements can inflict damage to an organism's genome, heightening rates of mutation.
Mutagens can be physical, such as radiation from UV rays, X-rays or extreme heat, or chemical (molecules that misplace base pairs or disrupt the helical shape of DNA).
Although mutations that cause changes in protein sequences can be harmful to an organism, on occasions the effect may be positive in a given environment.
[114] One possible explanation of the etiology of the relatively high frequency of CCR5-Δ32 in the European population is that it conferred resistance to the bubonic plague in mid-14th century Europe.
A newer theory suggests that the selective pressure on the CCR5 Delta 32 mutation was caused by smallpox instead of the bubonic plague.
[119] By introducing novel genetic qualities to a population of organisms, de novo mutations play a critical role in the combined forces of evolutionary change.
However, the weight of genetic diversity generated by mutational change is often considered a generally "weak" evolutionary force.
Since RNAs have relatively simpler composition than proteins, the structure of RNA molecules can be computationally predicted with high degree of accuracy.
[129] Lunzer et al.[130] tested the outcome of swapping divergent amino acids between two orthologous proteins of isopropymalate dehydrogenase (IMDH).
Corrigan et al. 2011 demonstrated how Staphylococcus aureus was able to grow normally without the presence of lipoteichoic acid due to compensatory mutations.
[133] Comas et al. 2012 used whole genome comparisons between clinical strains and lab derived mutants to determine the role and contribution of compensatory mutations in drug resistance to rifampicin.
[132] A similar study investigated the bacterial fitness associated with compensatory mutations in rifampin resistant Escherichia coli.
[134] Gong et al.[135] collected obtained genotype data of influenza nucleoprotein from different timelines and temporally ordered them according to their time of origin.
[140][134][133] In the human genome, the frequency and characteristics of de novo mutations have been studied as important contextual factors to our evolution.
[142] De novo mutations have also been researched as playing a crucial role in the persistence of genetic disease in humans.
With recents advancements in next-generation sequencing (NGS), all types of de novo mutations within the genome can be directly studied, the detection of which provides a magnitude of insight toward the causes of both rare and common genetic disorders.
The ability to conduct whole genome sequencing of parents and offspring allows for the comparison of mutation rates between generations, narrowing down the origin possibilities of certain genetic disorders.