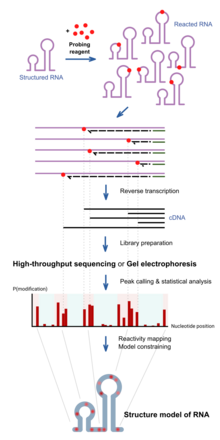
Nucleic acid structure determination
Experimental approaches of determining the structure of nucleic acids, such as RNA and DNA, can be largely classified into biophysical and biochemical methods.
Biophysical methods use the fundamental physical properties of molecules for structure determination, including X-ray crystallography, NMR and cryo-EM.
This is due to the greater degree of intrinsic disorder and dynamism in nucleic acid structures and the negatively charged (deoxy)ribose-phosphate backbones, which repel each other in close proximity.
Therefore, crystallized nucleic acids tend to be complexed with a protein of interest to provide structural order and neutralize the negative charge.
It has been especially useful in probing the structure of natural RNA oligonucleotides, which tend to adopt complex conformations such as stem-loops and pseudoknots.
One disadvantage, is that it is difficult to resolve nucleic acid or protein structures that are smaller than ~75 kilodaltons, partly due to the difficulty of having enough contrast to locate particles in this vitrified aqueous solution.
The collection of DNA molecules of various truncated lengths therefore informs the frequency of reaction at every base position, which reflects the structure profile along the RNA.
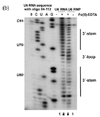
This is traditionally assayed by running the DNA on a gel, and the intensity of bands inform the frequency of observing a truncation at each position.
These can be detected when using high-throughput sequencing methods, and is sometimes employed for improved results of probing as mutational profiling (MaP).
Sites under protection from binding proteins or RNA tertiary structure would be cleaved by hydroxyl radical at a lower rate.
[17][18] Dimethyl sulfate, known as DMS, is a chemical that can be used to modify nucleic acids in order to determine secondary structure.
Improving upon truncation-based methods, DMS mutational profiling with sequencing (DMS-MaPseq) can detect multiple DMS modifications in a single RNA molecule, which enables one to obtain more information per read (for a read of 150 nt, typically two to three mutation sites, rather than zero to one truncation sites), determine structures of low-abundance RNAs, and identify subpopulations of RNAs with alternative secondary structures.
[21] Selective 2′-hydroxyl acylation analyzed by primer extension, or SHAPE, takes advantage of reagents that preferentially modify the backbone of RNA in structurally flexible regions.
Compared to the chemicals used in other RNA probing techniques, these reagents have the advantage of being largely unbiased to base identity, while remaining very sensitive to conformational dynamics.
Adduct formation is quantified for each nucleotide in a given RNA by extension of a complementary DNA primer with reverse transcriptase and comparison of the resulting fragments with those from an unmodified control.
[26] In SHAPE-Seq SHAPE is extended by bar-code based multiplexing combined with RNA-Seq and can be performed in a high-throughput fashion.
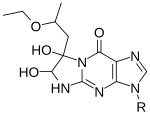
Kethoxal causes the modification of guanine, specifically altering the N1 and the exocyclic amino group (N2) simultaneously by covalent interaction.
Due to their chemical properties, these reagents can permeate readily into cells and can therefore be used to assay RNAs in their native cellular environments.
[34] Light-Activated Structural Examination of RNA (LASER) probing utilizes UV light to activate nicotinoyl azide (NAz), generating highly reactive nitrenium cation in water, which reacts with solvent accessible guanosine and adenosine of RNA at C-8 position through a barrierless Friedel-Crafts reaction.
This type of probing assay uses the structure dependent cleavage of RNA; single stranded regions are more flexible and unstable and will degrade over time.
The process of in-line probing involves incubation of structural or functional RNAs over a long period of time.
[38] In-line probing is a functional assay that can be used to determine structural changes in RNA in response to ligand binding.