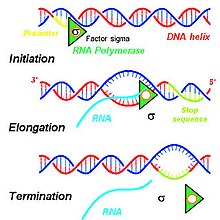
Primary transcript
All these steps involve a series of interactions to initiate and complete the transcription of DNA in the nucleus of eukaryotes.
Based on the needs of a given cell, certain DNA sequences are transcribed to produce a variety of RNA products to be translated into functional proteins for cellular use.
These DNA sequences bind to factors that contribute to the destabilization of the initiation complex required to activate RNA polymerase, and therefore inhibit transcription.
[6] Acetylation of histones induces repulsion between negative components within nucleosomes, allowing for RNA polymerase access.
Deacetylation of histones stabilizes tightly coiled nucleosomes, inhibiting RNA polymerase access.
RNA polymerase is often incapable of synthesizing a primary transcript if the targeted gene's promoter region contains specific methylated cytosines— residues that hinder binding of transcription-activating factors and recruit other enzymes to stabilize a tightly bound nucleosome structure, excluding access to RNA polymerase and preventing the production of primary transcripts.
An R-loop is a three-stranded nucleic acid structure containing a DNA-RNA hybrid region and an associated non-template single-stranded DNA.
Introns reduce R-loop formation and DNA damage in highly expressed yeast genes.
Even though these processes are tightly regulated and are usually accurate, occasionally they can make mistakes and leave behind DNA breaks that drive chromosomal rearrangements or cell death.
[9] Otherwise stated, the newly synthesized primary transcripts are modified in several ways to be converted to their mature, functional forms to produce different proteins and RNAs such as mRNA, tRNA, and rRNA.
[citation needed] The basic primary transcript modification process is similar for tRNA and rRNA in both eukaryotic and prokaryotic cells.
[9] For example, some prokaryotic bacterial mRNAs serve as templates for synthesis of proteins at the same time they are being produced via transcription.
Alternatively, pre-mRNA of eukaryotic cells undergo a wide range of modifications prior to their transport from the nucleus to cytoplasm where their mature forms are translated.
Furthermore, primary transcript processing provides a control for gene expression as well as a regulatory mechanism for the degradation rates of mRNAs.
In complex eukaryotic cells, one primary transcript is able to prepare large amounts of mature mRNAs due to alternative splicing.
[13] In HeLa cells, spliceosome groups on pre-mRNA were found to form within nuclear speckles, with this formation being temperature-dependent and influenced by specific RNA sequences.
The article entitled, "Alternative splicing of the human estrogen receptor alpha primary transcript: mechanisms of exon skipping" by Paola Ferro, Alessandra Forlani, Marco Muselli and Ulrich Pfeffer from the laboratory of Molecular Oncology at National Cancer Research Institute in Genoa, Italy, explains that 1785 nucleotides of the region in the DNA that codes for the estrogen receptor alpha (ER-alpha) are spread over a region that holds more than 300,000 nucleotides in the primary transcript.
Splicing of this pre-mRNA frequently leads to variants or different kinds of the mRNA lacking one or more exons or regions necessary for coding proteins.
This shows that the primary transcripts produced by these retroviruses do not always follow the normal path to protein production and convert back into DNA in order to multiply and expand.