Protein biosynthesis
During transcription, a section of DNA encoding a protein, known as a gene, is converted into a molecule called messenger RNA (mRNA).
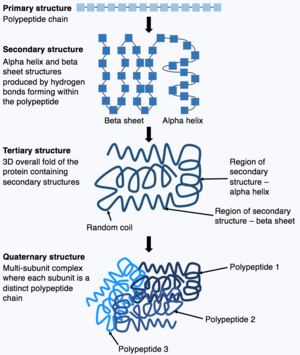
The ribosomes catalyze the formation of covalent peptide bonds between the encoded amino acids to form a polypeptide chain.
Alternatively, a mutation in the mRNA sequence changes the specific amino acid encoded at that position in the polypeptide chain.
However, in prokaryotes post-transcriptional modifications are not required so the mature mRNA molecule is immediately produced by transcription.
DNA has an antiparallel, double helix structure composed of two, complementary polynucleotide strands, held together by hydrogen bonds between the base pairs.
This property of directionality is due to the asymmetrical underlying nucleotide subunits, with a phosphate group on one side of the pentose sugar and a base on the other.
[1] The enzyme RNA polymerase binds to the exposed template strand and reads from the gene in the 3' to 5' direction.
The proofreading mechanisms allows the RNA polymerase to remove incorrect nucleotides (which are not complementary to the template strand of DNA) from the growing pre-mRNA molecule through an excision reaction.
Therefore, in the pre-mRNA molecule, all complementary bases which would be thymine in the coding DNA strand are replaced by uracil.
[1] In contrast, the 3' Poly(A) tail is added to the 3' end of the mRNA molecule and is composed of 100-200 adenine bases.
[6] During splicing, the intervening introns are removed from the pre-mRNA molecule by a multi-protein complex known as a spliceosome (composed of over 150 proteins and RNA).
[citation needed] During translation, ribosomes synthesize polypeptide chains from mRNA template molecules.
In eukaryotes, translation occurs in the cytoplasm of the cell, where the ribosomes are located either free floating or attached to the endoplasmic reticulum.
The ribosome reads the mRNA molecule in a 5'-3' direction and uses it as a template to determine the order of amino acids in the polypeptide chain.
[12] The ribosome initially attaches to the mRNA at the start codon (AUG) and begins to translate the molecule.
The correct tRNA with the anticodon (complementary 3 nucleotide sequence UAC) binds to the mRNA using the ribosome.
The ribosome then uses its peptidyl transferase enzymatic activity to catalyze the formation of the covalent peptide bond between the two adjacent amino acids.
This process continues with the ribosome moving along the mRNA molecule adding up to 15 amino acids per second to the polypeptide chain.
[6] Termination of the growing polypeptide chain occurs when the ribosome encounters a stop codon (UAA, UAG, or UGA) in the mRNA molecule.
[12] Dr. Har Gobind Khorana, a scientist originating from India, decoded the RNA sequences for about 20 amino acids.
[15] Through post-translational modifications, the diversity of proteins encoded by the genome is expanded by 2 to 3 orders of magnitude.
These proteases are often highly specific and cause hydrolysis of a limited number of peptide bonds within the target protein.
[17] Following translation, small chemical groups can be added onto amino acids within the mature protein structure.
[18] Examples of processes which add chemical groups to the target protein include methylation, acetylation and phosphorylation.
Phosphorylation is the reversible, covalent addition of a phosphate group to specific amino acids (serine, threonine and tyrosine) within the protein.
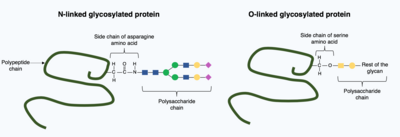
[16] In glycosylation, a polysaccharide molecule (known as a glycan) is covalently added to the target protein by glycosyltransferases enzymes and modified by glycosidases in the endoplasmic reticulum and Golgi apparatus.
Glycosylation can have a critical role in determining the final, folded 3D structure of the target protein.
In contrast, O-linked glycosylation is the sequential covalent addition of individual sugars onto the oxygen in the amino acids serine and threonine within the mature protein structure.
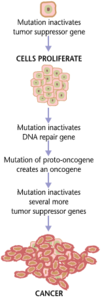
[22] Patients with sickle cell anemia have a missense or substitution mutation in the gene encoding the hemoglobin B subunit polypeptide chain.
In addition to cancer cells proliferating abnormally, they suppress the expression of anti-apoptotic or pro-apoptotic genes or proteins.