Reprogramming
In biology, reprogramming refers to erasure and remodeling of epigenetic marks, such as DNA methylation, during mammalian development or in cell culture.
Reprogrammings that are both large scale (10% to 100% of epigenetic marks) and rapid (hours to a few days) occur at three life stages of mammals.
Almost 100% of epigenetic marks are reprogrammed in two short periods early in development after fertilization of an ovum by a sperm.
In addition, almost 10% of DNA methylations in neurons of the hippocampus can be rapidly altered during formation of a strong fear memory.
After fertilization in mammals, DNA methylation patterns are largely erased and then re-established during early embryonic development.
Almost all of the methylations from the parents are erased, first during early embryogenesis, and again in gametogenesis, with demethylation and remethylation occurring each time.
Another period of rapid and almost complete demethylation occurs during gametogenesis within the primordial germ cells (PGCs).
[citation needed] After fertilization, the paternal chromosome is almost completely demethylated in six hours by an active process, before DNA replication (blue line in Figure).
The morula (at the 16 cell stage), has only a small amount of DNA methylation (black line in Figure).
At this point the PGC genomes display the lowest levels of DNA methylation of any cells in the entire life cycle [at embryonic day 13.5 (E13.5), see the second figure in this section].
Therefore, during the process of gametogenesis the primordial germ cells must have their original biparental DNA methylation patterns erased and re-established based on the sex of the transmitting parent.
After fertilization, the paternal and maternal genomes are demethylated in order to erase their epigenetic signatures and acquire totipotency.
[5] Despite the global nature of this process, there are certain sequences that avoid it, such as differentially methylated regions (DMRS) associated with imprinted genes, retrotransposons and centromeric heterochromatin.
[6] In vitro manipulation of pre-implantation embryos has been shown to disrupt methylation patterns at imprinted loci[7] and plays a crucial role in cloned animals.
[8] Learning and memory have levels of permanence, differing from other mental processes such as thought, language, and consciousness, which are temporary in nature.
Learning and memory can be either accumulated slowly (multiplication tables) or rapidly (touching a hot stove), but once attained, can be recalled into conscious use for a long time.
Rats subjected to one instance of contextual fear conditioning create an especially strong long-term memory.
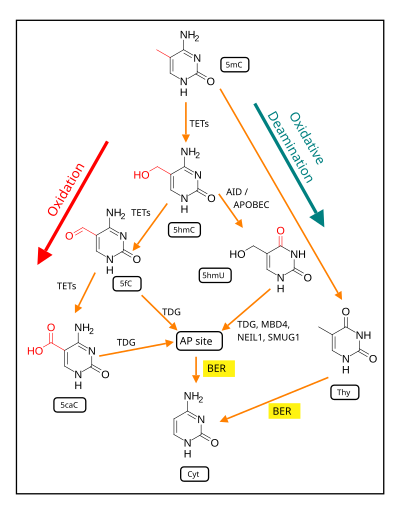
The Figure in this section indicates the central roles of ten-eleven translocation methylcytosine dioxygenases (TETs) in the demethylation of 5-methylcytosine to form cytosine.
The dominant TET1 isoform in most somatic tissues, at least in the mouse, arises from alternative promoter usage which gives rise to a short transcript and a truncated protein designated TET1s.
[16] The CXXC domain of the full-length TET3, which is the predominant form expressed in neurons, binds most strongly to CpGs where the C was converted to 5-carboxycytosine (5caC).
[25] This expression is linked to control of cognition, emotional response, social behavior and sensitivity to reward.
[26] The first person to successfully demonstrate reprogramming was John Gurdon, who in 1962 demonstrated that differentiated somatic cells could be reprogrammed back into an embryonic state when he managed to obtain swimming tadpoles following the transfer of differentiated intestinal epithelial cells into enucleated frog eggs.
This includes the expression of Oct-4 or Homeobox protein NANOG, while undergoing a mesenchymal–epithelial transition (MET), and the loss of apoptosis and senescence.
The use of microRNA and other small molecule-driven processes has been utilized as a means of increasing the efficiency of the differentiation from somatic cells to pluripotency.
[35] In the later stages of maturation, transgene silencing marks the start of the cell becoming independent from the induced transcription factor.
The use of microRNA, proteins, and different combinations of the OSKM factors have started to lead towards a higher efficiency rate of reprogramming.
It is carried out by the transfection of stem-cell associated genes into mature cells using viral vectors such as retroviruses.
The OSKM factors (Oct4, Sox2, Klf4, and c-Myc) were initially discovered by Yamanaka in 2006, by the induction of a mouse fibroblast into an induced pluripotent stem cell (iPSCs).
[32] Oct4 is part of the core regulatory genes needed for pluripotency, as it is seen in both embryonic stem cells and tumors.
[42] C/EBPα is considered a 'path breaker' to aid in preparing the cell for intake of the OSKM factors and specific transcription events.