Topologically associating domain
[2][3] Boundaries at both side of these domains are conserved between different mammalian cell types and even across species[2] and are highly enriched with CCCTC-binding factor (CTCF) and cohesin.
[7] Disruption of TAD boundaries are found to be associated with wide range of diseases such as cancer,[8][9][10] variety of limb malformations such as synpolydactyly, Cooks syndrome, and F-syndrome,[11] and number of brain disorders like Hypoplastic corpus callosum and Adult-onset demyelinating leukodystrophy.
[11] Furthermore, studies have revealed that interactions between promoters and enhancers spanning single or multiple TADs, are fundamental to the exact dynamics of gene expression.
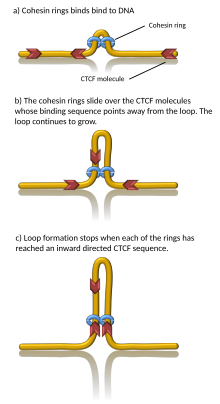
[4][5] Computer simulations have shown that chromatin loop extrusion driven by cohesin motors can generate TADs.
[27] Indeed, in vitro, cohesin has been observed to processively extrude DNA loops in an ATP-dependent manner[28][29][30] and stall at CTCF.
Removing a TAD boundary (for example, using CRISPR to delete the relevant region of the genome) can allow new promoter-enhancer contacts to form.
This can affect gene expression nearby - such misregulation has been shown to cause limb malformations (e.g. polydactyly) in humans and mice.
[47] Insulated neighborhoods, DNA loops formed by CTCF/cohesin-bound regions, are proposed to functionally underlie TADs.
[50] For example, genomic structural variants that disrupt TAD boundaries have been reported to cause developmental disorders such as human limb malformations.
[56] Lamina-associated domains (LADs) are parts of the chromatin that heavily interact with the lamina, a network-like structure at the inner membrane of the nucleus.