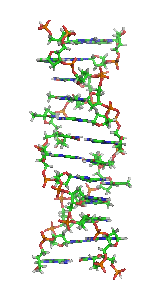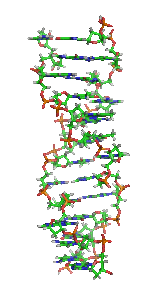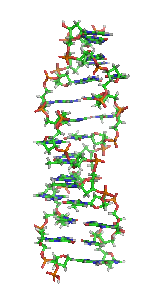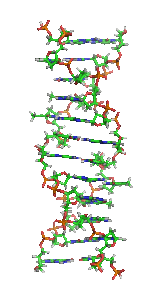
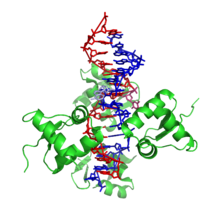
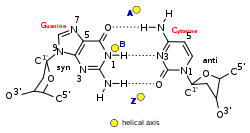
Z-DNA
It is a left-handed double helical structure in which the helix winds to the left in a zigzag pattern, instead of to the right, like the more common B-DNA form.
[1] They observed a "reverse" circular dichroism spectrum for such DNAs, and interpreted this incorrectly to mean that the strands wrapped around one another in a left-handed fashion.
Formation of this structure is generally unfavourable, although certain conditions can promote it; such as alternating purine–pyrimidine sequence (especially poly(dGC)2), negative DNA supercoiling or high salt and some cations (all at physiological temperature, 37 °C, and pH 7.3–7.4).
An algorithm for predicting the propensity of DNA to flip from the B-form to the Z-form, ZHunt, was written by P. Shing Ho in 1984 at MIT.
[10] This algorithm was later developed by Tracy Camp, P. Christoph Champ, Sandor Maurice, and Jeffrey M. Vargason for genome-wide mapping of Z-DNA (with Ho as the principal investigator).
[14] This was performed by measuring the intensity values between the donor and acceptor fluorescent dyes, also known as Fluorophores, in relation to each other as they exchange electrons, while tagged onto a DNA molecule.
Families with haploid ADAR transcriptome enabled mapping of Zα variants directly to disease, showing that genetic information is encoded in DNA by both shape and sequence.
[18] A role in regulating type I interferon responses in cancer is also supported by findings that 40% of a panel of tumors were dependent on the ADAR enzyme for survival.
[24] Developed behind the pathway of RNA polymerase through negative supercoiling, Z-DNA formed via active transcription has been shown to increase genetic instability, creating a propensity towards mutagenesis near promoters.
[26] In mammalian cells, the presence of such sequences was found to produce large genomic fragment deletions due to chromosomal double-strand breaks.
Both of these genetic modifications have been linked to the gene translocations found in cancers such as leukemia and lymphoma, since breakage regions in tumor cells have been plotted around Z-DNA-forming sequences.
Biological studies suggested that the Z-DNA binding domain of ADAR1 may localize this enzyme that modifies the sequence of the newly formed RNA to sites of active transcription.
[33][34] A role for Zα, Z-DNA and Z-RNA in defense of the genome against the invasion of Alu retro-elements in humans has evolved into a mechanism for the regulation of innate immune responses to dsRNA.
[35][18]Additionally, Zα domains are demonstrated to localize at the stress granules because of their innate ability in binding nucleic acid.
[37][38] Not only does the E3L protein have affinity to Z-DNA, it has also been found to play a role in the level of severity of virulence in mice caused by vaccinia virus, a type of poxvirus.
[37] Through research done by Kim, Y. et al. at the Massachusetts Institute of Technology, it was shown that replacing the N-terminus of the E3L protein with a Zα domain sequence, containing 14 Z-DNA binding residues similar to E3L, had little to no effect on pathogenicity of the virus in mice.
[36] Specifically, mutations in Tyr 48 and Pro 63 were found to reduce transactivation of the previously mentioned genes, as a result of loss of hydrogen bonding and london dispersion forces between E3L and the Z-DNA.