Formylation
In organic chemistry, the term is most commonly used with regards to aromatic compounds (for example the conversion of benzene to benzaldehyde in the Gattermann–Koch reaction).
Formylation reactions are a form of electrophilic aromatic substitution and therefore work best with electron-rich starting materials.
Benzene will react under aggressive conditions but deactivated rings such as pyridine are difficult to formylate effectively.
As it generally begins with nucleophilic attack by the aromatic group, the electron density of the ring is an important factor.
Methionine was first discovered to be formylated in E. coli by Marcker and Sanger in 1964[7] and was later identified to be involved in the initiation of protein synthesis in bacteria and organelles.
[9] This reaction is not used by eukaryotes or Archaea, as the presence of tRNAfMet in non bacterial cells is dubbed as intrusive material and quickly eliminated.
In E. coli, tRNAfMet is specifically recognized by initiation factor IF-2, as the formyl group blocks peptide bond formation at the N-terminus of methionine.
[9] Once protein synthesis is accomplished, the formyl group on methionine can be removed by peptide deformylase.
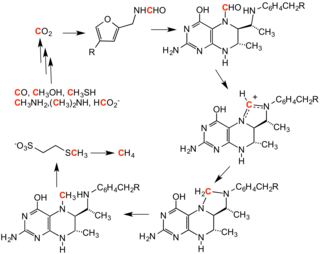
Two formylation reactions are required in the eleven step de novo synthesis of inosine monophosphate (IMP), the precursor of the purine ribonucleotides AMP and GMP.
A formyl phosphate intermediate has been detected in mutagenesis experiments, in which the mutant PurT GAR transforymylase had a weak affinity for formate.
[15] However, structural data indicates that the formate may be positioned for a direct attack on the γ-phosphate of ATP in the enzyme's active site to form the formylphosphate intermediate.
Histidine 268 is involved in deprotonation of the N5 nucleophile of AICAR, whereas Lysine 267 is proposed to stabilize the tetrahedral intermediate.
[13] ε-Formylation is one of many post-translational modifications that occur on histone proteins, which been shown to modulate chromatin conformations and gene activation.
Oxidative stress creates a significantly different environment in which acetyl-lysine can be quickly outcompeted by the formation of formyl-lysine due to the high reactivity of formylphosphate species.
A mechanism for the formation of formylphosphate has been proposed, which it is highly dependent on oxidatively damaged DNA and mainly driven by radical chemistry within the cell.
Oxidative stress is believed to play a role in the availability of lysine residues in the surface of proteins and the possibility of being formylated.
Inhibition of enzymes involved in purine biosynthesis has been exploited as a potential drug target for chemotherapy.
Cancer cells require high concentrations of purines to facilitate division[12] and tend to rely on de novo synthesis rather than the nucleotide salvage pathway.
[19] The first GAR transformylase inhibitor Lometrexol [(6R)5,10-dideazatetrahydrofolate] was developed in the 1980s through a collaboration between Eli Lilly and academic laboratories.
[19] Development of folate based inhibitors have been found to be particularly challenging as the inhibitors also down regulate the enzyme folypolyglutamate synthase, which adds additional γ-glutamates to monoglutamate folates and antifolates after entering the cell for increased enzyme affinity.
[21] Exome sequencing, has been used to identify a mutation in the gene coding for mitochondrial methionyl-tRNA formyltransferase (MTFMT) in patients with Leigh syndrome.