Metabolism
Lipids are usually defined as hydrophobic or amphipathic biological molecules but will dissolve in organic solvents such as ethanol, benzene or chloroform.
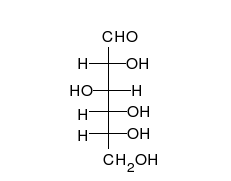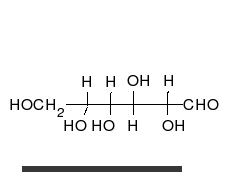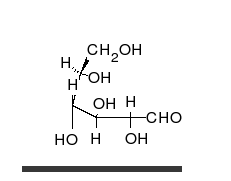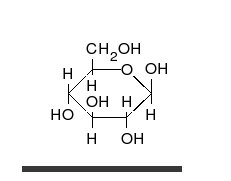
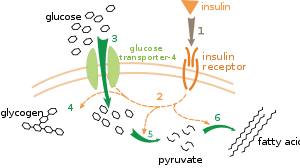
Carbohydrates are the most abundant biological molecules, and fill numerous roles, such as the storage and transport of energy (starch, glycogen) and structural components (cellulose in plants, chitin in animals).
[19] This common chemistry allows cells to use a small set of metabolic intermediates to carry chemical groups between different reactions.
[24] Inorganic elements play critical roles in metabolism; some are abundant (e.g. sodium and potassium) while others function at minute concentrations.
About 99% of a human's body weight is made up of the elements carbon, nitrogen, calcium, sodium, chlorine, potassium, hydrogen, phosphorus, oxygen and sulfur.
Organic compounds (proteins, lipids and carbohydrates) contain the majority of the carbon and nitrogen; most of the oxygen and hydrogen is present as water.
In the first stage, large organic molecules, such as proteins, polysaccharides or lipids, are digested into their smaller components outside cells.
Finally, the acetyl group on acetyl-CoA is oxidized to water and carbon dioxide in the citric acid cycle and electron transport chain, releasing more energy while reducing the coenzyme nicotinamide adenine dinucleotide (NAD+) into NADH.
[43] Pyruvate is an intermediate in several metabolic pathways, but the majority is converted to acetyl-CoA and fed into the citric acid cycle, which enables more ATP production by means of oxidative phosphorylation.
An alternative route for glucose breakdown is the pentose phosphate pathway, which produces less energy but supports anabolism (biomolecule synthesis).
This pathway reduces the coenzyme NADP+ to NADPH and produces pentose compounds such as ribose 5-phosphate for synthesis of many biomolecules such as nucleotides and aromatic amino acids.
M. tuberculosis can also grow on the lipid cholesterol as a sole source of carbon, and genes involved in the cholesterol-use pathway(s) have been validated as important during various stages of the infection lifecycle of M.
[49] In oxidative phosphorylation, the electrons removed from organic molecules in areas such as the citric acid cycle are transferred to oxygen and the energy released is used to make ATP.
The flow of protons makes the stalk subunit rotate, causing the active site of the synthase domain to change shape and phosphorylate adenosine diphosphate—turning it into ATP.
[62] In plants, algae, and cyanobacteria, photosystem II uses light energy to remove electrons from water, releasing oxygen as a waste product.
[citation needed] Anabolism is the set of constructive metabolic processes where the energy released by catabolism is used to synthesize complex molecules.
These differ by the route that carbon dioxide takes to the Calvin cycle, with C3 plants fixing CO2 directly, while C4 and CAM photosynthesis incorporate the CO2 into other compounds first, as adaptations to deal with intense sunlight and dry conditions.
This is important as it allows the formation and breakdown of glucose to be regulated separately, and prevents both pathways from running simultaneously in a futile cycle.
[91] This aminoacyl-tRNA is then a substrate for the ribosome, which joins the amino acid onto the elongating protein chain, using the sequence information in a messenger RNA.
[103] Here, processes including oxidative phosphorylation and the formation of disulfide bonds during protein folding produce reactive oxygen species such as hydrogen peroxide.
[3][120] This universal ancestral cell was prokaryotic and probably a methanogen that had extensive amino acid, nucleotide, carbohydrate and lipid metabolism.
[125] An alternative model comes from studies that trace the evolution of proteins' structures in metabolic networks, this has suggested that enzymes are pervasively recruited, borrowing enzymes to perform similar functions in different metabolic pathways (evident in the MANET database)[126] These recruitment processes result in an evolutionary enzymatic mosaic.
For example, in some parasites metabolic processes that are not essential for survival are lost and preformed amino acids, nucleotides and carbohydrates may instead be scavenged from the host.
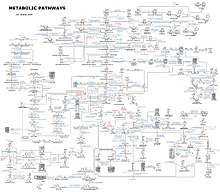
[132] An idea of the complexity of the metabolic networks in cells that contain thousands of different enzymes is given by the figure showing the interactions between just 43 proteins and 40 metabolites to the right: the sequences of genomes provide lists containing anything up to 26.500 genes.
[133] However, it is now possible to use this genomic data to reconstruct complete networks of biochemical reactions and produce more holistic mathematical models that may explain and predict their behavior.
[137][138] Bacterial metabolic networks are a striking example of bow-tie[139][140][141] organization, an architecture able to input a wide range of nutrients and produce a large variety of products and complex macromolecules using a relatively few intermediate common currencies.
Here, organisms such as yeast, plants or bacteria are genetically modified to make them more useful in biotechnology and aid the production of drugs such as antibiotics or industrial chemicals such as 1,3-propanediol and shikimic acid.
[148] Ibn al-Nafis described metabolism in his 1260 AD work titled Al-Risalah al-Kamiliyyah fil Siera al-Nabawiyyah (The Treatise of Kamil on the Prophet's Biography) which included the following phrase "Both the body and its parts are in a continuous state of dissolution and nourishment, so they are inevitably undergoing permanent change.
"[152] This discovery, along with the publication by Friedrich Wöhler in 1828 of a paper on the chemical synthesis of urea,[153] and is notable for being the first organic compound prepared from wholly inorganic precursors.
[156][157][75] Modern biochemical research has been greatly aided by the development of new techniques such as chromatography, X-ray diffraction, NMR spectroscopy, radioisotopic labelling, electron microscopy and molecular dynamics simulations.