Cofactor (biochemistry)
Cofactors can be classified into two types: inorganic ions and complex organic molecules called coenzymes.
[5][page needed] The International Union of Pure and Applied Chemistry (IUPAC) defines "coenzyme" a little differently, namely as a low-molecular-weight, non-protein organic compound that is loosely attached, participating in enzymatic reactions as a dissociable carrier of chemical groups or electrons; a prosthetic group is defined as a tightly bound, nonpolypeptide unit in a protein that is regenerated in each enzymatic turnover.
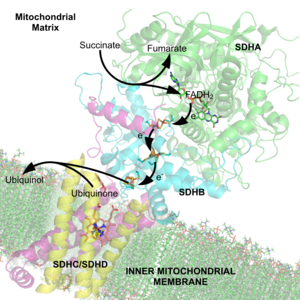
For example, the multienzyme complex pyruvate dehydrogenase[7] at the junction of glycolysis and the citric acid cycle requires five organic cofactors and one metal ion: loosely bound thiamine pyrophosphate (TPP), covalently bound lipoamide and flavin adenine dinucleotide (FAD), cosubstrates nicotinamide adenine dinucleotide (NAD+) and coenzyme A (CoA), and a metal ion (Mg2+).
It has been suggested that the AMP part of the molecule can be considered to be a kind of "handle" by which the enzyme can "grasp" the coenzyme to switch it between different catalytic centers.
On the other hand, "prosthetic group" emphasizes the nature of the binding of a cofactor to a protein (tight or covalent) and, thus, refers to a structural property.
In humans this list commonly includes iron, magnesium, manganese, cobalt, copper, zinc, and molybdenum.
[12] Although chromium deficiency causes impaired glucose tolerance, no human enzyme that uses this metal as a cofactor has been identified.
[17] Other organisms require additional metals as enzyme cofactors, such as vanadium in the nitrogenase of the nitrogen-fixing bacteria of the genus Azotobacter,[18] tungsten in the aldehyde ferredoxin oxidoreductase of the thermophilic archaean Pyrococcus furiosus,[19] and even cadmium in the carbonic anhydrase from the marine diatom Thalassiosira weissflogii.
One diverse set of examples is the heme proteins, which consist of a porphyrin ring coordinated to iron.
[29] Many organic cofactors also contain a nucleotide, such as the electron carriers NAD and FAD, and coenzyme A, which carries acyl groups.
[60] This common chemistry allows cells to use a small set of metabolic intermediates to carry chemical groups between different reactions.
[63] At least some of the current set of cofactors may, therefore, have been present in the last universal ancestor, which lived about 4 billion years ago.
[69] Similarly, pantetheine (a vitamin B5 derivative), a precursor of coenzyme A and thioester-dependent synthesis, can be formed spontaneously under evaporative conditions.
A computational method, IPRO, recently predicted mutations that experimentally switched the cofactor specificity of Candida boidinii xylose reductase from NADPH to NADH.
Through a long and difficult purification from yeast extracts, this heat-stable factor was identified as a nucleotide sugar phosphate by Hans von Euler-Chelpin.
[77] Other cofactors were identified throughout the early 20th century, with ATP being isolated in 1929 by Karl Lohmann,[78] and coenzyme A being discovered in 1945 by Fritz Albert Lipmann.
[80] This discovery was followed in the early 1940s by the work of Herman Kalckar, who established the link between the oxidation of sugars and the generation of ATP.
[82] Later, in 1949, Morris Friedkin and Albert L. Lehninger proved that NAD+ linked metabolic pathways such as the citric acid cycle and the synthesis of ATP.
[83] In a number of enzymes, the moiety that acts as a cofactor is formed by post-translational modification of a part of the protein sequence.
[84] These alterations are distinct from other post-translation protein modifications, such as phosphorylation, methylation, or glycosylation in that the amino acids typically acquire new functions.
[86] Characterization of protein-derived cofactors is conducted using X-ray crystallography and mass spectroscopy; structural data is necessary because sequencing does not readily identify the altered sites.
[87] In order to avoid confusion, it has been suggested that such proteins that have ligand-binding mediated activation or repression be referred to as coregulators.