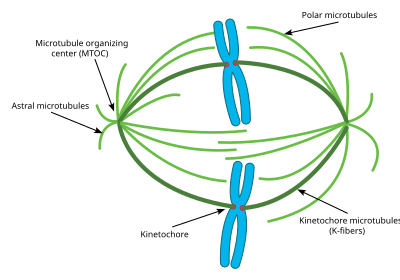
Spindle apparatus
Attachment of microtubules to chromosomes is mediated by kinetochores, which actively monitor spindle formation and prevent premature anaphase onset.
[13][14] Opposing the action of these microtubule-stabilizing proteins are a number of microtubule-depolymerizing factors which permit the dynamic remodeling of the mitotic spindle to promote chromosome congression and attainment of bipolarity.
The kinesin-13 superfamily of MAPs contains a class of plus-end-directed motor proteins with associated microtubule depolymerization activity including the well-studied mammalian MCAK and Xenopus XKCM1.
MCAK localizes to the growing tips of microtubules at kinetochores where it can trigger catastrophe in direct competition with stabilizing +TIP activity.
[15] These proteins harness the energy of ATP hydrolysis to induce destabilizing conformational changes in protofilament structure that cause kinesin release and microtubule depolymerization.
[15] Additional microtubule destabilizing proteins include Op18/stathmin and katanin which have roles in remodeling the mitotic spindle as well as promoting chromosome segregation during anaphase.
[17] The activities of these MAPs are carefully regulated to maintain proper microtubule dynamics during spindle assembly, with many of these proteins serving as Aurora and Polo-like kinase substrates.
The precise orientation of this complex is required to ensure accurate chromosome segregation and to specify the cell division plane.
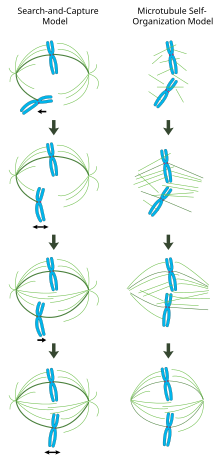
In contrast to the search-and-capture mechanism in which centrosomes largely dictate the organization of the mitotic spindle, this model proposes that microtubules are nucleated acentrosomally near chromosomes and spontaneously assemble into anti-parallel bundles and adopt a spindle-like structure.
[24] The guanine nucleotide exchange factor for the small GTPase Ran (Regulator of chromosome condensation 1 or RCC1) is attached to nucleosomes via core histones H2A and H2B.
[26] The gradient triggers release of spindle assembly factors (SAFs) from inhibitory interactions via the transport proteins importin β/α.
Mitotic entry triggers a dramatic reorganization of the duplicated genome, resulting in sister chromatids that are disentangled and separated from one another.
Condensation begins in prophase and chromosomes are maximally compacted into rod-shaped structures by the time they are aligned in the middle of the spindle at metaphase.
This gives mitotic chromosomes the classic "X" shape seen in karyotypes, with each condensed sister chromatid linked along their lengths by cohesin proteins and joined, often near the center, at the centromere.
[30][31][32] While these dynamic rearrangements are vitally important to ensure accurate and high-fidelity segregation of the genome, our understanding of mitotic chromosome structure remains largely incomplete.
A few specific molecular players have been identified, however: Topoisomerase II uses ATP hydrolysis to catalyze decatenation of DNA entanglements, promoting sister chromatid resolution.
[34] Experiments in Xenopus egg extracts have also implicated linker Histone H1 as an important regulator of mitotic chromosome compaction.