O-GlcNAc
[1] Due to the dynamic nature of O-GlcNAc and its presence on serine and threonine residues, O-GlcNAcylation is similar to protein phosphorylation in some respects.
Permeabilizing cells with detergent prior to radiolabeling greatly increased the amount of [3H]galactose incorporated into Galβ1-4GlcNAcitol, leading the authors to conclude that most of the O-linked GlcNAc monosaccharide residues were intracellular.
[16] The O-GlcNAc modification is removed by OGA in a hydrolysis mechanism involving anchimeric assistance (substrate-assisted catalysis) to yield the unmodified protein and GlcNAc.
For OGT, studies have shown that substrate recognition is regulated by a number of factors including aspartate[21] and asparagine[22] ladder motifs in the lumen of the superhelical TPR domain, active site residues,[23] and adaptor proteins.
For example, peracetylated GlcNAc (Ac4GlcNAz) is a cell-permeable azido sugar that is de-esterified intracellularly by esterases to GlcNAz and converted to UDP-GlcNAz in the hexosamine salvage pathway.
Ac4GalNAz shows enhanced labeling of O-GlcNAc versus Ac4GlcNAz, possibly due to a bottleneck in UDP-GlcNAc pyrophosphorylase processing of GlcNAz-1-P to UDP-GlcNAz.
[33] Ac3GlcN-β-Ala-NBD-α-1-P(Ac-SATE)2, a metabolic chemical reporter that is processed intracellularly to a fluorophore-labeled UDP-GlcNAc analogue, has been shown to achieve one-step fluorescent labeling of O-GlcNAc in live cells.
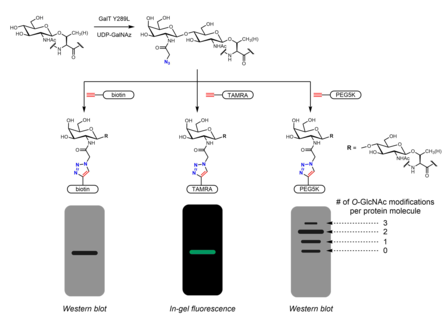
[30][46] The presence of GalNAz (and therefore also O-GlcNAc) can be detected with various alkyne-containing probes with identifiable tags such as biotin,[46] dye molecules,[30] and PEG.
[31] An engineered protein biosensor has been developed that can detect changes in O-GlcNAc levels using Förster resonance energy transfer.
As O-GlcNAc is substoichiometric and ion suppression occurs in the presence of unmodified peptides, an enrichment step is usually performed prior to mass spectrometry analysis.
[49] In order to facilitate site mapping with HCD, β-elimination followed by Michael addition with dithiothreitol (BEMAD) may be used to convert the labile O-GlcNAc modification into a more stable mass tag.
[56] The general procedure for this isotope-targeted glycoproteomics (IsoTaG) method is the following: Other methodologies have been developed for quantitative profiling of O-GlcNAc using differential isotopic labeling.
While serine/threonine phosphorylation may be modeled by mutagenesis to aspartate or glutamate, which have negatively charged carboxylate side chains, none of the 20 canonical amino acids sufficiently recapitulate the properties of O-GlcNAc.
[72] Recent studies have supported the use of S-GlcNAc as an enzymatically stable structural model of O-GlcNAc that can be incorporated through solid-phase peptide synthesis or site-directed mutagenesis.
O-GlcNAc is found in multiple locations on EZH2, the catalytic methyltransferase subunit of PRC2, and is thought to stabilize EZH2 prior to PRC2 complex formation and regulate di- and tri-methyltransferase activity.
OGT activity is in part regulated by UDP-GlcNAc concentration, making a link between cellular nutrient status and O-GlcNAc.
[128][129][130][131][132][133][134][135][136] Multiple companies have advanced OGA inhibitors into the clinic including Alectos Therapeutics, Asceneuron, Biogen, Eli Lilly, and Merck.
In this model, tau phosphorylation was not significantly affected by thiamet-G treatment, though decreased numbers of neurofibrillary tangles and slower motor neuron loss were observed.
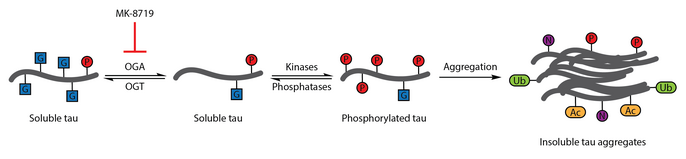
[96] OGA inhibition with MK-8719 is being investigated in clinical trials as a potential treatment strategy for Alzheimer's disease and other tauopathies including progressive supranuclear palsy.
Hypoxia induces PFK1 S529 O-GlcNAc and increases flux through the pentose phosphate pathway to generate more NADPH, which maintains glutathione levels and detoxifies reactive oxygen species, imparting a growth advantage to cancer cells.
[77][102] OGT stabilization of EZH2 in various breast cancer cell lines has been found to inhibit expression of tumor suppressor genes.
OGT knockdown or inhibition was found to downregulate the transcription factor FoxM1 and upregulate the cell-cycle inhibitor p27Kip1 (which is regulated by FoxM1-dependent expression of the E3 ubiquitin ligase component Skp2), causing G1 cell cycle arrest.
This appeared to be dependent on proteasomal degradation of FoxM1, as expression of a FoxM1 mutant lacking a degron rescued the effects of OGT knockdown.
Many components of the insulin signaling pathway, including β-catenin,[162] IR-β, IRS1, Akt, PDK1, and the p110α subunit of PI3K were found to be directly modified by O-GlcNAc.
[164] As PUGNAc also inhibits lysosomal β-hexosaminidases, the OGA-selective inhibitor NButGT was developed to further probe the relationship between O-GlcNAc and insulin signaling in 3T3-L1 adipocytes.
[165] Treatment of macrophages with lipopolysaccharide (LPS), a major component of the Gram-negative bacteria outer membrane, results in elevated O-GlcNAc in cellular and mouse models.
Suppressing O-GlcNAc with DON inhibited the O-GlcNAcylation and nuclear translocation of NF-κB, as well as downstream induction of inducible nitric oxide synthase and IL-1β production.
Specifically, O-GlcNAcylation of S430 on interferon regulatory factor-5 (IRF5) has been shown to promote its interaction with TNF receptor-associated factor 6 (TRAF6) in cellular and mouse models.
Analysis of clinical samples showed that blood glucose levels were elevated in IAV-infected patients compared to healthy individuals.
[167] Peptide therapeutics such as are attractive for their high specificity and potency, but they often have poor pharmacokinetic profiles due to their degradation by serum proteases.