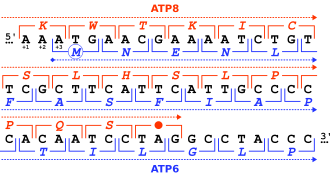
Overlapping gene
[4] In one model, the two proteins encoded by their respective overlapping genes evolve under similar selection pressures.
The proteins and the overlap region are highly conserved when strong selection against amino acid change is favored.
Overlapping genes are reasoned to evolve under strict constraints as a single nucleotide substitution is able to alter the structure and function of the two proteins simultaneously.
A study on the hepatitis B virus (HBV), whose DNA genome contains numerous overlapping genes, showed the mean number of synonymous nucleotide substitutions per site in overlapping coding regions was significantly lower than that of non-overlapping regions.
[15] The same study showed that it was possible for some of these overlapping regions and their proteins to diverge significantly from the original when there's weak selection against amino acid change.
The spacer domain of the polymerase and the pre-S1 region of a surface protein of HBV, for example, had a percentage of conserved amino acids of 30% and 40%, respectively.
[4] Overlapping genes are particularly common in rapidly evolving genomes, such as those of viruses, bacteria, and mitochondria.
[14][24] In 1977, Pierre-Paul Grassé proposed that one of the genes in the pair could have originated de novo by mutations to introduce novel ORFs in alternate reading frames; he described the mechanism as overprinting.
Which member of an overlapping gene pair is younger can be identified bioinformatically either by a more restricted phylogenetic distribution, or by less optimized codon usage.
[29][31] An alternative start site within the genome replication gene A of ΦX174 was shown to express a truncated protein with an identical coding sequence to the C-terminus of the original A protein but possessing a different function[32][33] It was concluded that other undiscovered sites of polypeptide synthesis could be hidden through the genome due to overlapping genes.
An identified de novo gene of another overlapping gene locus was shown to express a novel protein that induces lysis of E. coli by inhibiting biosynthesis of its cell wall[56], suggesting that de novo protein creation through the process of overprinting can be a significant factor in the evolution of pathogenicity of viruses.
Overprinted proteins often have unusual amino acid distributions and high levels of intrinsic disorder.
[28] However, examples of validated gene overlaps have been documented in a variety of eukaryotic organisms, including mammals such as mice and humans.
The retention and evolution of overlapping genes within viruses may also be due to capsid size limitations.
[2] Proteogenomic methods have been essential in discovering numerous overlapping genes and include a combination of techniques such as bottom-up proteomics, ribosome profiling, DNA sequencing, and perturbation.
It has been utilized to identify 180,000 alternate ORFs within previously annotated coding regions found in humans.
[69] Newly discovered ORFs such as these are verified using a variety of reverse genetics techniques, such as CRISPR-Cas9 and catalytically dead Cas9 (dCas9) disruption.