Permutation test
Under the null hypothesis, the distribution of the test statistic is obtained by calculating all possible values of the test statistic under possible rearrangements of the observed data.
The theory has evolved from the works of Ronald Fisher and E. J. G. Pitman in the 1930s.
[2] To illustrate the basic idea of a permutation test, suppose we collect random variables
The permutation test is designed to determine whether the observed difference between the sample means is large enough to reject, at some significance level, the null hypothesis H
First, the difference in means between the two samples is calculated: this is the observed value of the test statistic,
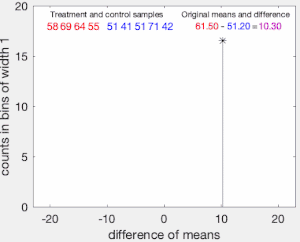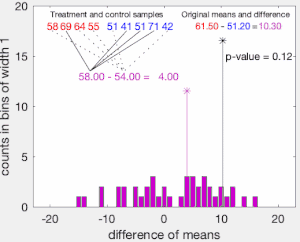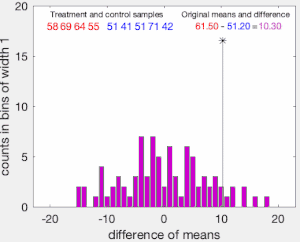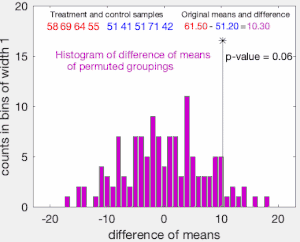
The set of these calculated differences is the exact distribution of possible differences (for this sample) under the null hypothesis that group labels are exchangeable (i.e., are randomly assigned).
The one-sided p-value of the test is calculated as the proportion of sampled permutations where the difference in means was greater than
The two-sided p-value of the test is calculated as the proportion of sampled permutations where the absolute difference was greater than
This is equivalent to performing a normal, unpaired permutation test, but restricting the set of valid permutations to only those which respect the paired nature of the data by forbidding both halves of any pair from being included in the same partition.
In the specific but common case where the test statistic is the mean, this is also equivalent to computing a single set of differences of each pair and iterating over all of the
Assuming that our experimental data come from data measured from two treatment groups, the method simply generates the distribution of mean differences under the assumption that the two groups are not distinct in terms of the measured variable.
When sample sizes are very large, the Pearson's chi-square test will give accurate results.
For small samples, the chi-square reference distribution cannot be assumed to give a correct description of the probability distribution of the test statistic, and in this situation the use of Fisher's exact test becomes more appropriate.
Thus one is always free to choose the statistic which best discriminates between hypothesis and alternative and which minimizes losses.
Permutation tests can be used for analyzing unbalanced designs[4] and for combining dependent tests on mixtures of categorical, ordinal, and metric data (Pesarin, 2001) [citation needed].
Permutation tests may be ideal for analyzing quantitized data that do not satisfy statistical assumptions underlying traditional parametric tests (e.g., t-tests, ANOVA),[5] see PERMANOVA.
Before the 1980s, the burden of creating the reference distribution was overwhelming except for data sets with small sample sizes.
Since the 1980s, the confluence of relatively inexpensive fast computers and the development of new sophisticated path algorithms applicable in special situations made the application of permutation test methods practical for a wide range of problems.
It also initiated the addition of exact-test options in the main statistical software packages and the appearance of specialized software for performing a wide range of uni- and multi-variable exact tests and computing test-based "exact" confidence intervals.
An important assumption behind a permutation test is that the observations are exchangeable under the null hypothesis.
This can be addressed in the same way the classic t-test has been extended to handle unequal variances: by employing the Welch statistic with Satterthwaite adjustment to the degrees of freedom.
In some cases, a permutation test based on a properly studentized statistic can be asymptotically exact even when the exchangeability assumption is violated.
An asymptotically equivalent permutation test can be created when there are too many possible orderings of the data to allow complete enumeration in a convenient manner.
This is done by generating the reference distribution by Monte Carlo sampling, which takes a small (relative to the total number of permutations) random sample of the possible replicates.
The earliest known references to this approach are Eden and Yates (1933) and Dwass (1957).
In the example above, the confidence interval only tells us that there is roughly a 50% chance that the p-value is smaller than 0.05, i.e. it is completely unclear whether the null hypothesis should be rejected at a level
can be established to be true or false with a very low probability of error.
or vice versa), the question of how many permutations to generate can be seen as the question of when to stop generating permutations, based on the outcomes of the simulations so far, in order to guarantee that the conclusion (which is either
Stopping rules to achieve this have been developed[12] which can be incorporated with minimal additional computational cost.
In fact, depending on the true underlying p-value it will often be found that the number of simulations required is remarkably small (e.g. as low as 5 and often not larger than 100) before a decision can be reached with virtual certainty.