S phase
[1] Since accurate duplication of the genome is critical to successful cell division, the processes that occur during S-phase are tightly regulated and widely conserved.
Entry into S-phase is controlled by the G1 restriction point (R), which commits cells to the remainder of the cell-cycle if there is adequate nutrients and growth signaling.
[2] This transition is essentially irreversible; after passing the restriction point, the cell will progress through S-phase even if environmental conditions become unfavorable.
[2] Accordingly, entry into S-phase is controlled by molecular pathways that facilitate a rapid, unidirectional shift in cell state.
[3] Active cyclin D-CDK4/6 complex induces release of E2F transcription factor, which in turn initiates expression of S-phase genes.
[3] Several E2F target genes promote further release of E2F, creating a positive feedback loop similar to the one found in yeast.
[7] SLBP binding is required for efficient processing, export, and translation of histone mRNAs, allowing it to function as a highly sensitive biochemical "switch".
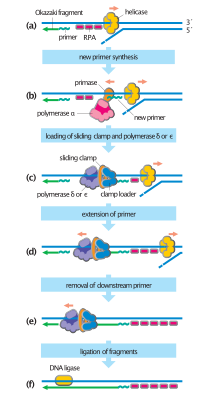
Translocation of MCM helicase along the leading strand disrupts parental nucleosome octamers, resulting in the release of H3-H4 and H2A-H2B subunits.
[10] However, for small domains approaching the size of individual genes, old nucleosomes are spread too thinly for accurate propagation of histone modifications.
Detection of DNA damage induces activation of three canonical S-phase "checkpoint pathways" that delay or arrest further cell cycle progression:[12] In addition to these canonical checkpoints, recent evidence suggests that abnormalities in histone supply and nucleosome assembly can also alter S-phase progression.
[14] This unique arrest phenotype is not associated with activation of canonical DNA damage pathways, indicating that nucleosome assembly and histone supply may be scrutinized by a novel S-phase checkpoint.