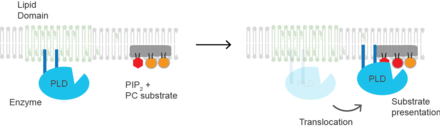
Substrate presentation
In the case of an interaction with an enzyme, the protein or organic substrate typically changes chemical form.
Substrate presentation differs from allosteric regulation in that the enzyme need not change its conformation to begin catalysis.
When cholesterol is low, the protein traffics to the disordered region and is cleaved by alpha secretase to produce a non-amylogenic product.
(ADAM17), also called TACE, is sequestered into lipid rafts away from its substrate, membrane bound tumor necrosis factor (mTNF).
[7] Cholesterol causes mTNF to cluster with ADAM17 in lipid rafts and shed soluble TNF (sTNF) which is an inflammatory cytokine.
When cells are loaded with cholesterol furin traffics to GM1 lipid rafts where it is localized with the palmitoylated spike protein of SARS-CoV-2 and primes it for viral entry.
[11][12] In low cholesterol ACE2 traffics the virus to TMPRSS2 which also cleaves and allows viral entry but through a putative surface mechanism that is much less efficient.
Within the plasma membrane, sequestration is primarily driven by packing of saturated lipid with cholesterol or phase separation at very small distances (< 100 nm).
For proteins that are both palmitoylated and bind PIP2, increasing the concentration of PIP2 favors trafficking of the enzyme out of lipid rafts to PIP2.
While speculative, the profound effect of cholesterol and PUFAs on human health is likely through physiological regulation of lipid raft function in cells.
Mechanical force (shear or swell) can independently disrupt the packing and resultant affinity of palmitate to lipid rafts.
[16] The mechanosensitive ion channel TREK-1 is released from cholesterol dependent lipid rafts in response to mechanical force.