Topoisomerase
The first type II topoisomerase to be discovered was DNA gyrase from bacteria, by Martin Gellert and coworkers in 1976,[5] and also characterized by Nicholas Cozzarelli and co-workers.
The exception among the type I topoisomerases, reverse gyrase, which contains a helicase domain (EC 3.6.4.12) and introduces positive supercoiling in an ATP-dependent manner.
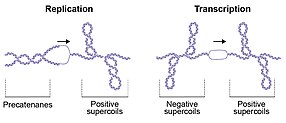
The double-helical structure of DNA involves the intertwining of the two polynucleotide strands around each other, which potentially gives rise to topological problems.
[14][15] Closed-circular double-stranded DNA can be described by 3 parameters: Linking number (Lk), Twist (Tw) and Writhe (Wr) (Fig.
[17][18] Lk cannot be altered without breaking one or both strands of the helix; Tw and Wr are interconvertible and depend upon the solution conditions.
If the positive supercoils are not relaxed, progression of the replication fork is impeded, whereas failure to unlink the daughter strands prevents genome segregation, which is required for cell division.
[21] These topological perturbations must be resolved for DNA metabolism to proceed, allowing the cell to efficiently replicate, transcribe and partition the genome to enable cellular division and vitality.
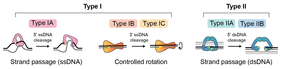
For strand passage to occur, topo IA must undergo a conformational change to open the DNA gate and allow T-segment transfer.
Reverse gyrase, which occurs in thermophilic archaea, comprises a type IA topo coupled to a helicase, and is the only known enzyme that can introduce positive supercoils into DNA.
[27] This controlled-rotation mechanism was first described for Vaccinia topo I[27][28] and permits DNA rotation of the free end around the intact strand, the speed being controlled by 'friction' within the enzyme cavity, before the nick is re-ligated (Fig.
The G segment is part of a much longer piece of DNA (>100 bp) that is wrapped around the enzyme, one arm of which forms the T-segment that is passed through the double-stranded break (Fig.
In the case of gyrase, a substantial amount of the free energy from ATP hydrolysis is transduced into torsional stress in DNA, i.e. supercoiling is an energy-requiring process.
6), such as novobiocin, clorobiocin and coumermycin A1, are natural products from Streptomyces that inhibit the ATPase reaction of gyrase and topo IV.
[37] Although they can be very potent against their target, they suffer from permeability and toxicity issues, and thus have not enjoyed the level of clinical success of the FQs.
There are a number of protein inhibitors of gyrase, including the bacterial toxins CcdB, MccB17, and ParE,[38][39][40] that stabilize the cleavage complex, in a similar manner to FQs.
[32][33][44][45][46][47] Most of these compounds act in a similar way to FQs, i.e. by stabilizing the DNA-protein covalent cleavage complex; for this they have become known as topoisomerase poisons, distinct from catalytic inhibitors.
[33] Camptothecin and its derivatives act by stabilizing the topo I cleavage complex, preventing religation of the protein-mediated nick in the DNA.
Although CPT derivatives stabilize a single-strand cleavage complex, subsequent collisions with replication or transcription machinery are thought to generate toxic double-stranded DNA breaks.
These compounds are used as first or second line therapies to treat cancers including colorectal, ovarian, lung, breast, and cervical.
7) and its close relative teniposide (VM-26) are epipodophyllotoxin derivatives obtained from the rhizome of wild mandrake that target topo II by stabilizing the covalent cleavage complex and preventing religation of the cleaved DNA.
Etoposide treatment can result in secondary leukemias arising from specific genomic translocations, mainly involving topo IIβ.
7) and the related derivatives daunorubicin, epirubicin, and idarubicin are anthracyclines obtained from the bacterium Streptomyces[48] that target human topo II, stabilizing the cleavage complex in a similar manner to other topoisomerase poisons.
[33] Additional cytotoxicity stems from redox reactions involving anthracyclines that generate reactive oxygen species.
[52][53][54][55] Topo IIβ, with other associated enzymes,[54] appears to be important for the release of paused RNA polymerase at highly transcribed or long genes.
[56][57][58] Stimulus-induced DNA double-strand breaks (DSBs) that are limited to a short-term (10 minutes to 2 hours) are induced by topo IIβ in the promoter regions of signal-regulated genes.
These DSBs allow rapid up-regulation of expression of such signal responsive genes in a number of systems (see Table below).
When the induced DNA double-strand break has been repaired, then transcription of the signal-responsive gene returns to a low basal level.
[52] Topo IIβ and PARP-1 were found to be constitutively present at a moderate level near the transcription start site of a promoter of a signal-responsive gene.
After the signal occurred, topo IIβ caused a double-strand break and PARP-1 was involved in replacing histone H1 by HMGB1/HMGA2, which can promote transcription.
This assemblage was all present at the linker DNA adjacent to a single nucleosome in the promoter region of a gene (see Figure).