Ubiquitin
These 'linking' residues are represented by a "K" or "M" (the one-letter amino acid notation of lysine and methionine, respectively) and a number, referring to its position in the ubiquitin molecule as in K48, K29 or M1.
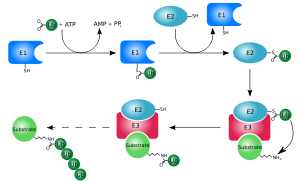
The first ubiquitin molecule is covalently bound through its C-terminal carboxylate group to a particular lysine, cysteine, serine, threonine or N-terminus of the target protein.
The basic functions of ubiquitin and the components of the ubiquitylation pathway were elucidated in the early 1980s at the Technion by Aaron Ciechanover, Avram Hershko, and Irwin Rose for which the Nobel Prize in Chemistry was awarded in 2004.
A heat-stable polypeptide present in these extracts, ATP-dependent proteolysis factor 1 (APF-1), was found to become covalently attached to the model protein substrate lysozyme in an ATP- and Mg2+-dependent process.
Multi-ubiquitin chains at least four ubiquitin molecules long must be attached to a lysine residue on the condemned protein in order for it to be recognised by the 26S proteasome.
Although initially believed to target proteins for proteasomal degradation,[72] linear ubiquitin later proved to be indispensable for NF-kB signaling.
[8][11][75] The ubiquitylation system functions in a wide variety of cellular processes, including:[78] Multi-monoubiquitylation can mark transmembrane proteins (for example, receptors) for removal from membranes (internalisation) and fulfil several signalling roles within the cell.
When DNA is damaged by ultra-violet radiation or chemicals, the SUMO molecule that is attached to a lysine residue is replaced by ubiquitin.
[86][87] Deubiquitinating enzymes (deubiquitinases; DUBs) oppose the role of ubiquitylation by removing ubiquitin from substrate proteins.
[94] Higher levels of ubiquilin in the brain have been shown to decrease malformation of amyloid precursor protein (APP), which plays a key role in triggering Alzheimer's disease.
Ubiquitin and ubiquitin-like molecules extensively regulate immune signal transduction pathways at virtually all stages, including steady-state repression, activation during infection, and attenuation upon clearance.
[96] The retinoic acid-inducible gene I (RIG-I) protein is a primary immune system sensor for viral and other invasive RNA in human cells.
[98] Immunohistochemistry using antibodies to ubiquitin can identify abnormal accumulations of this protein inside cells, indicating a disease process.
These protein accumulations are referred to as inclusion bodies (which is a general term for any microscopically visible collection of abnormal material in a cell).
First evidence of the importance of the ubiquitin/proteasome pathway in oncogenic processes was observed due to the high antitumor activity of proteasome inhibitors.
VHL complex targets a member of the hypoxia-inducible transcription factor family (HIF) for degradation by interacting with the oxygen-dependent destruction domain under normoxic conditions.
HIF activates downstream targets such as the vascular endothelial growth factor (VEGF), promoting angiogenesis.
[114] Oncogenic types of the human papillomavirus (HPV) are known to hijack cellular ubiquitin-proteasome pathway for viral infection and replication.
[112][115] Efp, or estrogen-inducible RING-finger protein, is an E3 ubiquitin ligase whose overexpression has been shown to be the major cause of estrogen-independent breast cancer.
[108][115] As the most aggressive cancer originated in the brain, mutations found in patients with glioblastoma are related to the deletion of a part of the extracellular domain of the epidermal growth factor receptor (EGFR).
[116] Finding a specific molecule that selectively inhibits the activity of a certain E3 ligase and/or the protein–protein interactions implicated in the disease remains as one of the important and expanding research area.
Moreover, as ubiquitination is a multi-step process with various players and intermediate forms, consideration of the much complex interactions between components needs to be taken heavily into account while designing the small molecule inhibitors.
It is also believed that the Saccharomyces cerevisiae protein Urm1, a ubiquitin-related modifier, is a "molecular fossil" that connects the evolutionary relation with the prokaryotic ubiquitin-like molecules and ubiquitin.
[128] Archaea have a functionally closer homolog of the ubiquitin modification system, where "sampylation" with SAMPs (small archaeal modifier proteins) is performed.
[130] Prokaryotic ubiquitin-like protein (Pup) is a functional analog of ubiquitin which has been found in the gram-positive bacterial phylum Actinomycetota.
It serves the same function (targeting proteins for degradations), although the enzymology of ubiquitylation and pupylation is different, and the two families share no homology.
Bacteria encode evolutionary predecessors of cGAS, called cGAS/DncV-like nucleotidyltransferases (CD-NTases), which detect bacteriophage infection and produce diverse nucleotide second messengers.
Recent research has shown that CD-NTase-associated protein 2 (Cap2) primes bacterial CD-NTases for activation through a ubiquitin transferase-like mechanism.
This research demonstrates that bacteria control immune signaling using an ancient, minimized ubiquitin transferase-like system, providing insights into the evolution of E1 and E2 machinery across domains of life.
ANUBL1; BAG1; BAT3/BAG6; C1orf131; DDI1; DDI2; FAU; HERPUD1; HERPUD2; HOPS; IKBKB; ISG15; LOC391257; MIDN; NEDD8; OASL; PARK2; RAD23A; RAD23B; RPS27A; SACS; 8U SF3A1; SUMO1; SUMO2; SUMO3; SUMO4; TMUB1; TMUB2; UBA52; UBB; UBC; UBD; UBFD1; UBL4A; UBL4B; UBL7; UBLCP1; UBQLN1; UBQLN2; UBQLN3; UBQLN4; UBQLNL; UBTD1; UBTD2; UHRF1; UHRF2; Currently available prediction programs are: