Carbonic anhydrase
[6] Carbonic anhydrase was initially isolated and characterised from red blood cells in 1933, with simultaneous reports by Meldrum and Roughton (at Cambridge University in the United Kingdom) and by Stadie and O’Brien (at the University of Pennsylvania in the United States),[7][8] both while searching for a "catalytic factor... necessary for rapid transit of the HCO3− [bicarbonate anion] from the erythrocyte to... pulmonary capillar[ies]".
[10] Carbonic anhydrase is a very ancient enzyme found in both domains of prokaryotes that exists in six different classes among most of the living organisms.
[citation needed] An enzyme is a substance that acts as a catalyst in living organisms which helps to speed up chemical reactions.
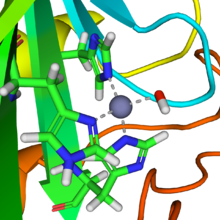
In mammalian CA II, the active site consists of the following: a hard Lewis acid Zn+2 metal atom coordinated to His -94, -96, and -119 residues 109˚ apart from one another and a hydroxide ion (pKa=6.8; 120° in Td configuration, a hydrophobic pocket adjacent to Zinc-bound hydroxide consisting of by Val-143 at its base and Val-121, Trp-209, and Leu-198 at its neck, a Proton Shuttling Residue (PSR) His-64 H+ shuttles H+ in and out of active site via conformational switching, and a hydrogen bonding network consisting of Thr-199 hydroxyl group and Glu-106 the carboxyl group which stabilizes the Zinc-bound hydroxide by facilitating the orientation of water molecules in the active side to a specific geometric configuration.
Carbon dioxide is transported in the blood in three forms: A zinc prosthetic group in the enzyme is coordinated in three positions by histidine side-chains.
A fourth histidine is close to the water ligand, facilitating formation of Zn-OH center, which binds CO2 to give a zinc bicarbonate.
The active site also features a pocket suited for carbon dioxide, bringing it close to the hydroxide group.
Here the Zn2+ acts as a Lewis acid that lowers the pKa of the coordinated OH2 ligand from ~7-8 down to 6.8 as Td , which drives the deprotonation of water to a hydroxide ion and the free proton is neutralized by the surrounding buffer.
In step 4), the coordinated water ligand is deprotonated facilitated by the Zn+2 to generate another hydroxide ion to start the cycle over again.
The CA enzymes found in mammals are divided into four broad subgroups,[citation needed] which, in turn consist of several homologous classes of genes: There are three additional "acatalytic" human carbonic anhydrase isoforms (CA-VIII, CA-X, and CA-XI) (CA8, CA10, CA11) whose functions remain unclear.
Two signature patterns for this family have been identified: The gamma class of CAs comes from methanogens, methane-producing archaea that grow in hot springs.
The zeta class of CAs occurs exclusively in bacteria in a few chemolithotrophs and marine cyanobacteria that contain cso-carboxysomes.
[28] Recent 3-dimensional analyses[27] suggest that ζ-CA bears some structural resemblance to β-CA, particularly near the metal ion site.
These are a group of enzymes previously thought to belong to the alpha family of CAs, however it has been demonstrated that η-CAs have unique features, such as their metal ion coordination pattern.
[30] In diatoms, the ι-CA is essential for the CO2-concentrating mechanisms and - in contrast to other CA classes - it can use manganese instead of zinc as metal cofactor.
In the best-studied α-carbonic anhydrase form present in animals, the zinc ion is coordinated by the imidazole rings of 3 histidine residues, His94, His96, and His119.
Plants contain a different form called β-carbonic anhydrase, which, from an evolutionary standpoint, is a distinct enzyme, but participates in the same reaction and also uses a zinc ion in its active site.
In plants, carbonic anhydrase helps raise the concentration of CO2 within the chloroplast in order to increase the carboxylation rate of the enzyme RuBisCO.
T. weissflogii, a species of phytoplankton common to many marine ecosystems, was found to contain carbonic anhydrase with a cadmium ion in place of zinc.
[34] Although the concentration of cadmium in sea water is also low (about 1x10−16 molar), there is an environmental advantage to being able to use either metal depending on which is more available at the time.
[36] Additionally, like the other carbonic anhydrases, CDCA makes the reaction go almost as fast as the diffusion rate of its substrates, and it can be inhibited by sulfonamide and sulfamate derivatives.
The active site of CDCA is essentially "gated" by a chain of nine amino acids with glycine residues at positions 1 and 9.
[36] As a borderline acid, zinc will not bind as tightly to the cysteine ligands as cadmium would, but the enzyme will still be active and reasonably efficient.
[38] A pilot run with the more stable CA on a flue stream that consisted of 12–13% mol composition CO₂ had a capture rate of 63.6% over a 60-hour period with no noticeable effects in enzyme performance.
CA was placed in a N-methyldiethanolamine (MDEA) solution where it served to increase the concentration difference (driving force) of CO2 between the flue stream of the power plant and liquid phase in a liquid-gas contactor.