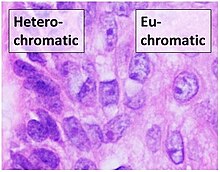
Chromatin
Regions of DNA containing genes which are actively transcribed ("turned on") are less tightly compacted and closely associated with RNA polymerases in a structure known as euchromatin, while regions containing inactive genes ("turned off") are generally more condensed and associated with structural proteins in heterochromatin.
[2] The consequences in terms of chromatin accessibility and compaction depend both on the modified amino acid and the type of modification.
For example, it was proposed that a bivalent structure (with trimethylation of both lysine 4 and 27 on histone H3) is involved in early mammalian development.
A- and B-DNA are very similar, forming right-handed helices, whereas Z-DNA is a left-handed helix with a zig-zag phosphate backbone.
These play a dual role of a site of recognition by many proteins and as a sink for torsional stress from RNA polymerase or nucleosome binding.
The order and sequences of these chemical structures of DNA are reflected as information available for the creation and control of human organisms.
Sugar and phosphate molecules are also paired with these bases, making DNA nucleotides arrange 2 long spiral strands unitedly called “double helix”.
Nucleosomes, with about 20 to 60 base pairs of linker DNA, can form, under non-physiological conditions, an approximately 11 nm beads on a string fibre.
This is due primarily to the varying physical properties of different DNA sequences: For instance, adenine (A), and thymine (T) is more favorably compressed into the inner minor grooves.
The existing models commonly accept that the nucleosomes lie perpendicular to the axis of the fibre, with linker histones arranged internally.
[2] Chromatin and its interaction with enzymes has been researched, and a conclusion being made is that it is relevant and an important factor in gene expression.
Vincent G. Allfrey, a professor at Rockefeller University, stated that RNA synthesis is related to histone acetylation.
Specifically, RNA polymerase and transcriptional proteins have been shown to congregate into droplets via phase separation, and recent studies have suggested that 10 nm chromatin demonstrates liquid-like behavior increasing the targetability of genomic DNA.
[25] The interactions between linker histones and disordered tail regions act as an electrostatic glue organizing large-scale chromatin into a dynamic, liquid-like domain.
[2] The phenomenon, as opposed to simple probabilistic models of transcription, can account for the high variability in gene expression occurring between cells in isogenic populations.
[27] It is proposed that in yeast, regions devoid of histones become very fragile after transcription; HMO1, an HMG-box protein, helps in stabilizing nucleosomes-free chromatin.
Many factors influence how the repair route is selected, including the cell cycle phase and chromatin segment where the break occurred.
In order to maintain genomic integrity, “homologous recombination and classical non-homologous end joining process” has been followed by DNA to be repaired.
[32] The packaging of eukaryotic DNA into chromatin presents a barrier to all DNA-based processes that require recruitment of enzymes to their sites of action.
[36] γH2AX, the phosphorylated form of H2AX is also involved in the early steps leading to chromatin decondensation after DNA damage occurrence.
[37] γH2AX (H2AX phosphorylated on serine 139) can be detected as soon as 20 seconds after irradiation of cells (with DNA double-strand break formation), and half maximum accumulation of γH2AX occurs in one minute.
[38] RNF8 mediates extensive chromatin decondensation, through its subsequent interaction with CHD4,[39] a component of the nucleosome remodeling and deacetylase complex NuRD.
It has been shown that the process of chromatin-loop extrusion is ideally suited to actively unknot chromatin fibres in interphase chromosomes.
[49] The term, introduced by Walther Flemming, has multiple meanings: The first definition allows for "chromatins" to be defined in other domains of life like bacteria and archaea, using any DNA-binding proteins that condenses the molecule.





Left: 1 start helix "solenoid" structure.
Right: 2 start loose helix structure.
Note: the histones are omitted in this diagram - only the DNA is shown.

Linker DNA in yellow and nucleosomal DNA in pink.