DNA oxidation
The one electron reduction potentials of the nucleosides (in volts versus NHE) are guanine 1.29, adenine 1.42, cytosine 1.6 and thymine 1.7.
Swenberg et al.[9] measured average amounts of steady state endogenous DNA damages in mammalian cells.
As reviewed by Valavanidis et al.[11] increased levels of 8-oxo-dG in a tissue can serve as a biomarker of oxidative stress.
[15] Li et al.[16] reviewed studies indicating that one or more BER proteins also participate(s) in epigenetic alterations involving DNA methylation, demethylation or reactions coupled to histone modification.
Nishida et al.[17] examined 8-oxo-dG levels and also evaluated promoter methylation of 11 tumor suppressor genes (TSGs) in 128 liver biopsy samples.
Yasui et al.[18] examined the fate of 8-oxo-dG when this oxidized derivative of deoxyguanosine was inserted into the thymidine kinase gene in a chromosome within human lymphoblastoid cells in culture.
As reviewed by Wang et al.,[19] oxidized guanine appears to have multiple regulatory roles in gene expression.
As noted by Wang et al.,[19] genes prone to be actively transcribed are densely distributed in high GC-content regions of the genome.
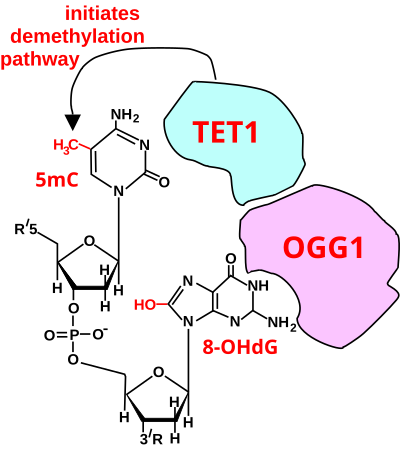
The experimental basis establishing this mode was also reviewed by Seifermann and Epe[20] A second mode of gene regulation by DNA oxidation at a guanine,[19][21] occurs when an 8-oxo-dG is formed in a guanine rich, potential G-quadruplex-forming sequence (PQS) in the coding strand of a promoter, after which active OGG1 excises the 8-oxo-dG and generates an apurinic/apyrimidinic site (AP site).
The AP site enables melting of the duplex to unmask the PQS, adopting a G-quadruplex fold (G4 structure/motif) that has a regulatory role in transcription activation.
Chromodomain helicase DNA-binding protein 4 (CHD4), a component of the (NuRD) complex, is recruited by OGG1 to oxidative DNA damage sites.
Perillo et al.,[22][23] showed that the lysine-specific histone demethylase LSD1 generates a local burst of reactive oxygen species (ROS) that induces oxidation of nearby nucleotides when carrying out its function.
The oxidation of DNA by LSD1 in the course of the demethylation of histone H3 at lysine 9 was shown to be required for the recruitment of OGG1 and also topoisomerase IIβ to the promoter region of bcl-2, an estrogen-responsive gene, and subsequent transcription initiation.
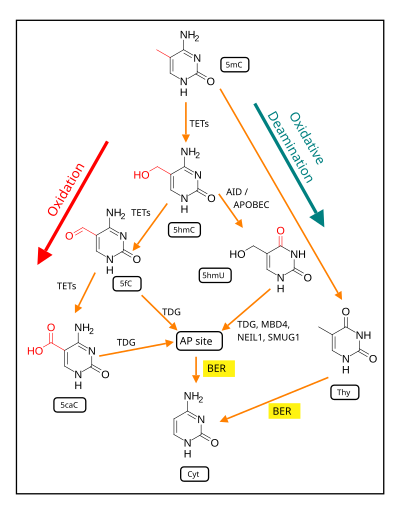
As indicated below, the first step in de-methylation of methylated cytosine at a CpG site is oxidation of the guanine to form 8-oxo-dG.
[32] Evidence that oxidative stress induced DNA damage plays a role in bipolar disorder has been reviewed by Raza et al.[33] Bipolar patients have elevated levels of oxidatively induced DNA base damages even during periods of stable mental state.
[35] Postmortem studies of elderly patients with chronic schizophrenia showed that oxidative DNA damage is increased in the hippocampus region of the brain.
[36] The mean proportion of neurons with the oxidized DNA base 8-oxo-dG was 10-fold higher in patients with schizophrenia than in comparison individuals.
The level of oxidative stress that a cell endures is reflected by the quantity of reactive oxygen species (ROS).
Actually, monitoring a system by applying the isotopical label [18O]-H2O2 shows greater oxidation in cellular RNA than in DNA.
Oxidation randomly damages RNAs, and each attack bring problems to the normal cellular metabolism.
Although alteration of genetic information on mRNA is relatively rare, oxidation on mRNAs in vitro and in vivo results in low translation efficiency and aberrant protein products.
Besides its abundance, 8-hydroxydeoxyguanosine (8-oxodG) and 8-hydroxyguanosine (8-oxoG) are identified as the most detrimental oxidation lesions for their mutagenic effect,[45] in which this non-canonical counterpart can faultily pair with both adenine and cytosine at the same efficiency.
So far, approaches developed to directly measure 8-oxoG level include HPLC-based analysis and assays employing monoclonal anti-8-oxoG antibody.
[50][51] Some potential factors include ribonucleases, which are suspected to selectively degrade damaged RNAs under stresses.