DNA damage (naturally occurring)
DNA damages cause changes in the structure of the genetic material and prevents the replication mechanism from functioning and performing properly.
[12] Other types of endogeneous DNA damages, given below with their frequencies of occurrence, include depurinations, depyrimidinations, double-strand breaks, O6-methylguanines, and cytosine deamination.
[13] The list below shows some frequencies with which new naturally occurring DNA damages arise per day, due to endogenous cellular processes.
Swenberg et al.[30] measured average amounts of selected steady state endogenous DNA damages in mammalian cells.
On the other hand, the ability to trigger apoptosis in the presence of excess un-repaired DNA damage is critical for prevention of cancer.
[45] Bile acids, stored in the gall bladder, are released into the small intestine in response to fat in the diet.
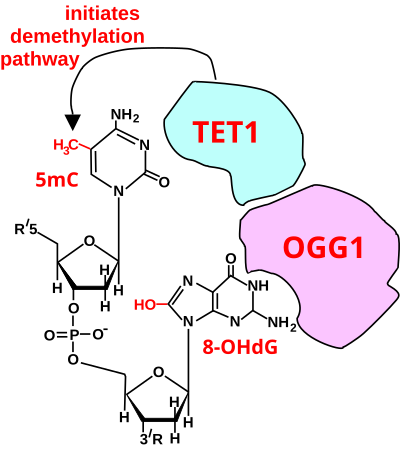
[61] Oxidized bases in DNA are produced in cells treated with Hoechst dye followed by micro-irradiation with 405 nm light.
Multiple DSBs can be induced by irradiating sensitized cells (labeled with 5'-bromo-2'-deoxyuridine and with Hoechst dye) with 780 nm light.
[70] γH2AX (H2AX phosphorylated on serine 139) can be detected as soon as 20 seconds after irradiation of cells (with DNA double-strand break formation), and half maximum accumulation of γH2AX occurs in one minute.
[71] RNF8 mediates extensive chromatin decondensation, through its subsequent interaction with CHD4,[72] a component of the nucleosome remodeling and deacetylase complex NuRD.
This is followed by phosphorylation of the cell cycle checkpoint protein Chk1, initiating its function, about 10 minutes after DNA is damaged.
The AP site enables melting of the duplex to unmask the PQS, adopting a G-quadruplex fold (G4 structure/motif) that has a regulatory role in transcription activation.
Chromodomain helicase DNA-binding protein 4 (CHD4), a component of the (NuRD) complex, is recruited by OGG1 to oxidative DNA damage sites.
[84] Double-stranded breaks (DSBs) in regions of DNA related to neuronal activity are produced by a variety of mechanisms within and around the genome.
The enzyme topoisomerase II, or TOPIIβ plays a key role in DSB formation by aiding in the demethylation or loosening of histones wrapped around the double helix to promote transcription.
[85] Once the chromatin structure is opened, DSBs are more likely to accumulate, however, this is normally repaired by TOPIIβ through its intrinsic religation ability that rejoins the cleaved DNA ends.
[85][86] Rapid expression of egr-1, c-Fos, and Arc IEGs have been observed in response to increased neuronal activity in the hippocampus region of the brain where memory processing takes place.
Changes in level of activity have been used in studies as a biomarker to trace the overlap between DSBs and increased histone H3K4 methylation in promoter regions of IEGs.
[85][88] Other studies have indicated that transposable elements (TEs) can cause DSBs through endogenous activity that involves using endonuclease enzymes to insert and cleave target DNA at random sites.
[citation needed] Buildup of DSBs more broadly leads to the degeneration of neurons, hindering the function of memory and learning processes.
Due to their lack of cell division and high metabolic activity, neurons are especially prone to DNA damage.
Environmental factors such as viruses and a high-fat diet have also been associated with disrupted function of DNA repair molecules.
[citation needed] One targeted therapy for treating patients with AD has involved suppression of the BRCA1 gene in human brains, initially tested in transgenic mice, where DSB levels were observed to have increased and memory loss had occurred, suggesting that BRCA1 could "serve as a therapeutic target for AD and AD-related dementia.
For example, in bacteria, a regulatory network aimed at repairing DNA damages (called the SOS response in Escherichia coli) has been found in many bacterial species.
For example, in fission yeast and humans, RecA homologues promote duplex-duplex DNA-strand exchange needed for repair of many types of DNA lesions.
In a non-replicating cell, a single-strand break or other type of damage in the transcribed strand of DNA can block RNA polymerase II-catalysed transcription.
Various species of animals exhibit similar mechanisms of cellular delay in response to DNA damage, which can be caused by exposure to x-irradiation.
The budding yeast Saccharomyces cerevisiae has specifically been studied because progression through the cell cycle can be followed via nuclear morphology with ease.
[citation needed] Through extensive experiments, researchers have been able to illuminate the role that the RAD genes play in delaying cell division in response to DNA damage.
[citation needed] Although the function of RAD9 has primarily been studied in the budding yeast Saccharomyces cerevisiae, many of the cell cycle control mechanisms are similar between species.