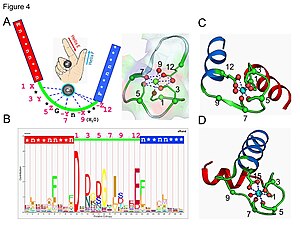
EF hand
The EF-hand motif contains a helix–loop–helix topology, much like the spread thumb and forefinger of the human hand, in which the Ca2+ ions are coordinated by ligands within the loop.
The EF-hand consists of two alpha helices linked by a short loop region (usually about 12 amino acids) that usually binds calcium ions.
Five of the loop residues bind calcium and thus have a strong preference for oxygen-containing side chains, especially aspartate and glutamate.
Upon binding to Ca2+, this motif may undergo conformational changes that enable Ca2+-regulated functions as seen in Ca2+ effectors such as calmodulin (CaM) and troponin C (TnC) and Ca2+ buffers such as calreticulin and calbindin D9k.
[2] This high selectivity is due to the relatively rigid coordination geometry, the presence of multiple charged amino acid side chains in the binding site, as well as the ion solvation properties.
[3][4][5] Pattern (motif signature) search is one of the most straightforward ways to predict continuous EF-hand Ca2+-binding sites in proteins.
Functionally, the EF-hands can be divided into two classes: The first group is the largest and includes the most well-known members of the family such as calmodulin, troponin C and S100B.