Protein isoform
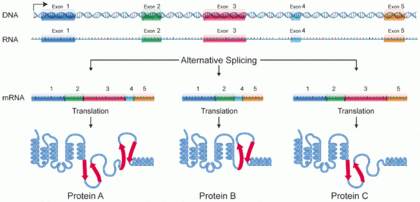
[3] One single gene has the ability to produce multiple proteins that differ both in structure and composition;[4][5] this process is regulated by the alternative splicing of mRNA, though it is not clear to what extent such a process affects the diversity of the human proteome, as the abundance of mRNA transcript isoforms does not necessarily correlate with the abundance of protein isoforms.
[9] Alternative splicing is the main post-transcriptional modification process that produces mRNA transcript isoforms, and is a major molecular mechanism that may contribute to protein diversity.
[5] The spliceosome, a large ribonucleoprotein, is the molecular machine inside the nucleus responsible for RNA cleavage and ligation, removing non-protein coding segments (introns).
[12] Consequently, although alternative splicing has been implicated as an important link between variation and disease, there is no conclusive evidence that it acts primarily by producing novel protein isoforms.
Generally, one protein isoform is labeled as the canonical sequence based on criteria such as its prevalence and similarity to orthologous—or functionally analogous—sequences in other species.
However, some isoforms show much greater divergence (for example, through trans-splicing), and can share few to no exons with the canonical sequence.
Typical examples of glycoproteins consisting of glycoforms are the blood proteins as orosomucoid, antitrypsin, and haptoglobin.
An unusual glycoform variation is seen in neuronal cell adhesion molecule, NCAM involving polysialic acids, PSA.