microRNA
[1] In 2024, American scientists Victor Ambros and Gary Ruvkun were awarded the Nobel Prize in Physiology or Medicine for their work on the discovery of miRNA and its role in post-transcriptional gene regulation.
However, additional insight into its mode of action required simultaneously published work by Gary Ruvkun's team, including Wightman and Ha.
For example: Pre-miRNAs, pri-miRNAs and genes that lead to 100% identical mature miRNAs but that are located at different places in the genome are indicated with an additional dash-number suffix.
The resulting transcript is capped with a specially modified nucleotide at the 5' end, polyadenylated with multiple adenosines (a poly(A) tail),[56][52] and spliced.
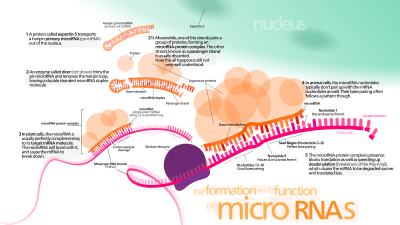
This protein, a member of the karyopherin family, recognizes a two-nucleotide overhang left by the RNase III enzyme Drosha at the 3' end of the pre-miRNA hairpin.
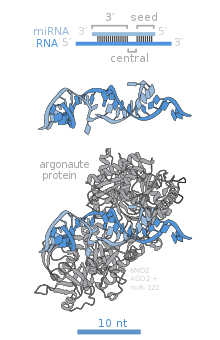
The duplex is then transported out of the nucleus to the cytoplasm by a protein called Hasty (HST), an Exportin 5 homolog, where they disassemble and the mature miRNA is incorporated into the RISC.
Some miRNAs work as buffers of random gene expression changes arising due to stochastic events in transcription, translation and protein stability.
As indicated by work in the model organism Arabidopsis thaliana (thale cress), mature plant miRNAs appear to be stabilized by the addition of methyl moieties at the 3' end.
[101][102] miRNAs occasionally also cause histone modification and DNA methylation of promoter sites, which affects the expression of target genes.
[114][115][116][117][118] While core components of the microRNA pathway are conserved between plants and animals, miRNA repertoires in the two kingdoms appear to have emerged independently with different primary modes of action.
[121] microRNAs' origin as a regulatory mechanism developed from previous RNAi machinery that was initially used as a defense against exogenous genetic material such as viruses.
MicroRNAs play a vital role in the regulation of gene expression in all non-ctenophore animals investigated thus far except for Trichoplax adhaerens, the first known member of the phylum Placozoa.
[135] While researchers focused on miRNA expression in physiological and pathological processes, various technical variables related to microRNA isolation emerged.
The locked conformation of LNA results in enhanced hybridization properties and increases sensitivity and selectivity, making it ideal for detection of short miRNA.
[150][151] While this is usually done after miRNAs of interest have been detected (e. g. because of high expression levels), ideas for analysis tools that integrate mRNA- and miRNA-expression information have been proposed.
[163] In-vitro studies suggested that miR-205 and miR-373 may functionally induce different features of mucinous-associated neoplastic progression in intestinal epithelial cells.
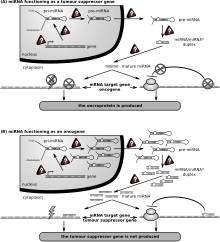
[173] However, altered expression of microRNAs, causing DNA repair deficiencies, are frequently associated with cancers and may be an important causal factor.
Among 68 sporadic colon cancers with reduced expression of the DNA mismatch repair protein MLH1, most were found to be deficient due to epigenetic methylation of the CpG island of the MLH1 gene.
Human neoplasias, including thyroid, prostatic, cervical, colorectal, pancreatic and ovarian carcinomas, show a strong increase of HMGA1a and HMGA1b proteins.
[178] Transgenic mice with HMGA1 targeted to lymphoid cells develop aggressive lymphoma, showing that high HMGA1 expression is associated with cancers and that HMGA1 can act as an oncogene.
[181] Single Nucleotide polymorphisms (SNPs) can alter the binding of miRNAs on 3'UTRs for example the case of hsa-mir181a and hsa-mir181b on the CDON tumor suppressor gene.
[195] Coinciding with these results, miR-712 is also upregulated in endothelial cells exposed to naturally occurring d-flow in the greater curvature of the aortic arch.
The obstruction of the blood flow means the brain cannot receive necessary nutrients, such as oxygen and glucose, and remove wastes, such as carbon dioxide.
[213] miRNA global regulation of many downstream genes deems significant regarding the reorganization or synaptic connections or long term neural adaptations involving the behavioral change from alcohol consumption to withdrawal and/or dependence.
[216] miR-155, important in regulating alcohol-induced neuroinflammation responses, was found to be upregulated, suggesting the role of microglia and inflammatory cytokines in alcohol pathophysiology.
[217] Downregulation of miR-382 was found in the nucleus accumbens, a structure in the basal forebrain significant in regulating feelings of reward that power motivational habits.
[218] Alternatively, overexpressing miR-382 resulted in attenuated drinking and the inhibition of DRD1 and delta fosB upregulation in rat models of alcoholism, demonstrating the possibility of using miRNA-targeted pharmaceuticals in treatments.
Conversely, ectopic expression of the miRNAs 155, 221, and 222 significantly inhibited adipogenesis and repressed induction of the master regulators PPARγ and CCAAT/enhancer-binding protein alpha (CEBPA).
[222] When let-7 was ectopically overexpressed to mimic accelerated aging, mice became insulin-resistant, and thus more prone to high fat diet-induced obesity and diabetes.
[223] In contrast when let-7 was inhibited by injections of let-7-specific antagomirs, mice become more insulin-sensitive and remarkably resistant to high fat diet-induced obesity and diabetes.