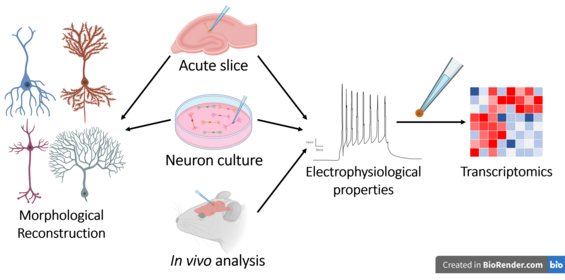
Patch-sequencing
Patch-sequencing (patch-seq) is a modification of patch-clamp technique that combines electrophysiological, transcriptomic and morphological characterization of individual neurons.
Disordered geometry and projection patterns has been linked to a diverse set of psychiatric and neurological conditions including autism and schizophrenia though the behavioral relevance of these phenotypes is not yet understood.
Previous attempts at neuronal classification by morpho-electric properties have been limited by the use of incompatible methodologies and different cell line selection.
[3] With the advent of single-cell RNA-sequencing (scRNA-seq) it was hoped that there would exist genes that would be consistently expressed only in neurons with specific classically defined properties.
However it appeared that scRNA-seq only served to reinforce the fact that overly rigid cell type definitions are not always the best way to characterize neurons.
[4] Furthermore gene expression is dynamically regulated, varying over various time scales in response to activity in cell type specific ways to allow for neuronal plasticity.
Linking the sequencing information to a neuronal subtype, defined previously by electrophysiological and morphological characteristics is a slow and complicated process.
[12] Cell culture preparation is the easiest to apply patch clamp to and give the experimenter control over what ligands the neuron is exposed to, for instance hormones or neurotransmitters.
Like wise complex post-hoc processing of transcriptomic data is often required as well in order to handle a large number of confounds when collecting cytosolic contents from cells via the pipette.
[22] The patch pipette is designed for whole cell recording so its opening diameter is larger than experiments done to examine single ion-channels.
For the most part standard patch clamp protocols may be used although there are some small situation dependent modifications to the pipette and the internal solution.
[23] Negative pressure is applied to enhance the seal which will be better for recording as well as prevent intra-cellular fluid leakage and contamination after or during collection of cell contents for sequencing.
[18] Once a seal has been established cells are subject to different stimulation regimes using the voltage clamp, such as ramps, square pulses and noisy current injections.
[10] The addition of RNase inhibitors, such as EGTA, enhances transcriptome analysis by preserving higher quality RNA from the samples.
[9] Recording time can take between 1 and 15 minutes without affecting the neuron structure due to swelling, with lower values increasing throughput of the technique.
It is crucial to have healthy acute or live brain slices for electrophysiological recordings, as the health of the neuron significantly impacts the quality of the data obtained.
Healthy brain slices are typically prepared using tools such as the Compresstome vibratome, ensuring optimal conditions for accurate and reliable recordings.
Larger variations in neuronal size compared to rodent models means greater variability in the amount of negative pressure needed to be applied to extract the cytosol and nucleus.
[14] Designing workflow for processing and combining the resulting multimodal data depends on the particular research question patch-seq is being applied to.
[36] Including morphological data has proven to be challenging as it is a computer vision task, a notoriously complicated problem in machine learning.
Neuroscientists have applied this technique to a variety of experiments and protocols to discover new cell subtypes based on correlations between transcriptomic data and neuronal morpho-electric properties.
[10] The technique is particularly well suited for neuroscience but in general any tissue where it is of interest to simultaneous know electrophysiology, morphology, and transcriptomics would find use for patch-seq.
[10] Nevertheless no other electrophysiological recording's technique can match patch-seq's temporal resolution or ability to characterize specific ionic currents nor offer the capacity to produce a voltage clamp.
For these reasons automating the technique is an area of active investigation by many laboratories around the world for applications using patch clamping including patch-seq.
[14] The proper integration of the massive number of images with high quantities of noise, and the structural disruption caused by nucleus extraction, makes it a computational challenge in neuroscience.
Automated patch clamping can either be done blind without imaging information instead relying on pressure sensors to provide input for algorithms guiding robotic rigs to form seals and record.