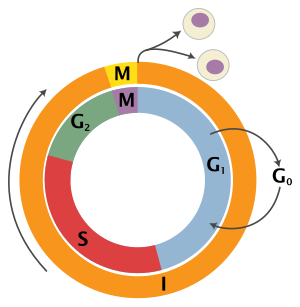
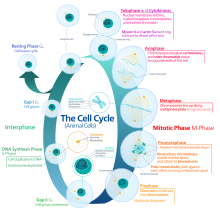
Cell cycle
Interphase is a series of changes that takes place in a newly formed cell and its nucleus before it becomes capable of division again.
[8] During the process of mitosis the pairs of chromosomes condense and attach to microtubules that pull the sister chromatids to opposite sides of the cell.
For example, animal cells undergo an "open" mitosis, where the nuclear envelope breaks down before the chromosomes separate, while fungi such as Aspergillus nidulans and Saccharomyces cerevisiae (yeast) undergo a "closed" mitosis, where chromosomes divide within an intact cell nucleus.
Even in animals, cytokinesis and mitosis may occur independently, for instance during certain stages of fruit fly embryonic development.
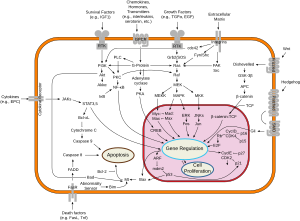
[11] Errors in mitosis can result in cell death through apoptosis or cause mutations that may lead to cancer.
[12] Leland H. Hartwell, R. Timothy Hunt, and Paul M. Nurse won the 2001 Nobel Prize in Physiology or Medicine for their discovery of these central molecules.
[13] Many of the genes encoding cyclins and CDKs are conserved among all eukaryotes, but in general, more complex organisms have more elaborate cell cycle control systems that incorporate more individual components.
Results from a study of E2F transcriptional dynamics at the single-cell level argue that the role of G1 cyclin-CDK activities, in particular cyclin D-CDK4/6, is to tune the timing rather than the commitment of cell cycle entry.
The reason for prevention of gaps in replication is fairly clear, because daughter cells that are missing all or part of crucial genes will die.
The un-phosphorylated Rb tumour suppressor functions in inducing cell cycle exit and maintaining G0 arrest (senescence).
[23] Hyperphosphorylated Rb is completely dissociated from E2F, enabling further expression of a wide range of E2F target genes are required for driving cells to proceed into S phase [1].
The hyperphosphorylated Rb dissociates from the E2F/DP1/Rb complex (which was bound to the E2F responsive genes, effectively "blocking" them from transcription), activating E2F.
Genes that regulate the amplitude of E2F accumulation, such as Myc, determine the commitment in cell cycle and S phase entry.
[30] Given the observations of cyclin D-Cdk 4/6 functions, inhibition of Cdk 4/6 should result in preventing a malignant tumor from proliferating.
Three Cdk4/6 inhibitors – palbociclib, ribociclib, and abemaciclib – currently received FDA approval for clinical use to treat advanced-stage or metastatic, hormone-receptor-positive (HR-positive, HR+), HER2-negative (HER2-) breast cancer.
Current evidence suggests that a semi-autonomous transcriptional network acts in concert with the CDK-cyclin machinery to regulate the cell cycle.
[37] Genome-wide studies using high throughput technologies have identified the transcription factors that bind to the promoters of yeast genes, and correlating these findings with temporal expression patterns have allowed the identification of transcription factors that drive phase-specific gene expression.
[35][39] Experimental evidence also suggests that gene expression can oscillate with the period seen in dividing wild-type cells independently of the CDK machinery.
Before the midblastula transition, zygotic transcription does not occur and all needed proteins, such as the B-type cyclins, are translated from maternally loaded mRNA.
[43] This confirms previous predictions from mathematical modeling of a global causal coordination between DNA replication origin activity and mRNA expression,[44][45][46] and shows that mathematical modeling of DNA microarray data can be used to correctly predict previously unknown biological modes of regulation.
[47] Checkpoints prevent cell cycle progression at specific points, allowing verification of necessary phase processes and repair of DNA damage.
Many types of cancer are caused by mutations that allow the cells to speed through the various checkpoints or even skip them altogether.
An alternative model of the cell cycle response to DNA damage has also been proposed, known as the postreplication checkpoint.
Among other things, this induces the now fertilized oocyte to return from its previously dormant, G0, state back into the cell cycle and on to mitotic replication and division.
Note, these fusions are fragments that contain a nuclear localization signal and ubiquitination sites for degradation, but are not functional proteins.
Sulfhydryls are natural substances that protect cells from radiation damage and tend to be at their highest levels in S and at their lowest near mitosis.
[55] Non-homologous end joining, a less accurate and more mutagenic process for repairing double strand breaks, is active throughout the cell cycle.
A central component of the cell cycle is its ability to coordinate the continuous and periodic duplications of different cellular elements, which evolved with the formation of the genome.
[56] Cell-cycle progression is controlled by the oscillating concentrations of different cyclins and the resulting molecular interactions from the various cyclin-dependent kinases (CDKs).
The sum of these regulatory networks creates a hysteretic and bistable scheme, despite the specific proteins being highly diverged.