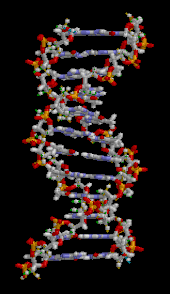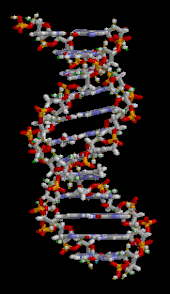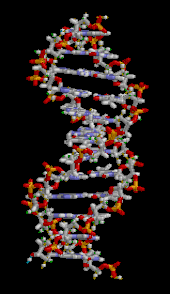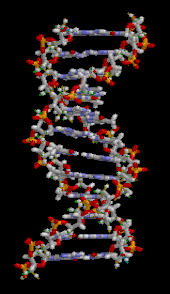
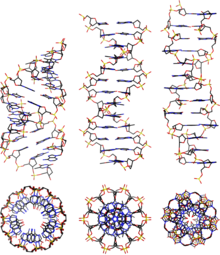
DNA
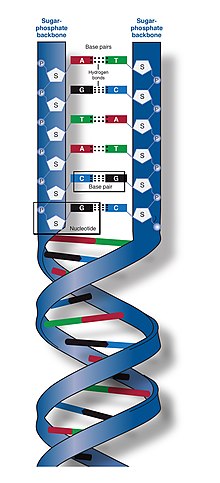
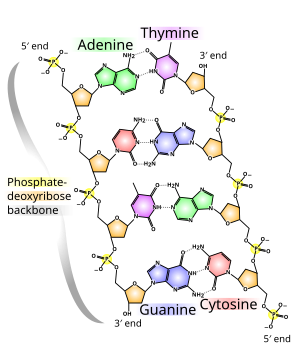
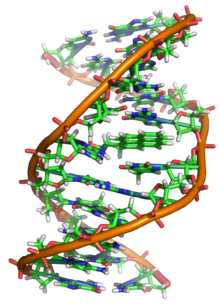
As a result, proteins such as transcription factors that can bind to specific sequences in double-stranded DNA usually make contact with the sides of the bases exposed in the major groove.
[28] In biology, parts of the DNA double helix that need to separate easily, such as the TATAAT Pribnow box in some promoters, tend to have a high AT content, making the strands easier to pull apart.
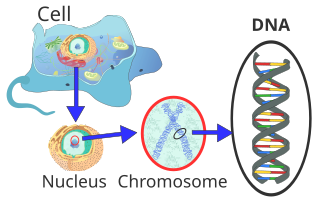
[30] In humans, the total female diploid nuclear genome per cell extends for 6.37 Gigabase pairs (Gbp), is 208.23 cm long and weighs 6.51 picograms (pg).
[46] The first published reports of A-DNA X-ray diffraction patterns—and also B-DNA—used analyses based on Patterson functions that provided only a limited amount of structural information for oriented fibers of DNA.
[47][48] An alternative analysis was proposed by Wilkins et al. in 1953 for the in vivo B-DNA X-ray diffraction-scattering patterns of highly hydrated DNA fibers in terms of squares of Bessel functions.
[49] In the same journal, James Watson and Francis Crick presented their molecular modeling analysis of the DNA X-ray diffraction patterns to suggest that the structure was a double helix.
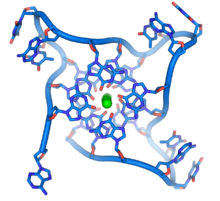
[65] These guanine-rich sequences may stabilize chromosome ends by forming structures of stacked sets of four-base units, rather than the usual base pairs found in other DNA molecules.
[83] On the other hand, oxidants such as free radicals or hydrogen peroxide produce multiple forms of damage, including base modifications, particularly of guanosine, and double-strand breaks.
[85] Of these oxidative lesions, the most dangerous are double-strand breaks, as these are difficult to repair and can produce point mutations, insertions, deletions from the DNA sequence, and chromosomal translocations.
[106] Various possible functions have been proposed for eDNA: it may be involved in horizontal gene transfer;[107] it may provide nutrients;[108] and it may act as a buffer to recruit or titrate ions or antibiotics.
[113] Under the name of environmental DNA eDNA has seen increased use in the natural sciences as a survey tool for ecology, monitoring the movements and presence of species in water, air, or on land, and assessing an area's biodiversity.
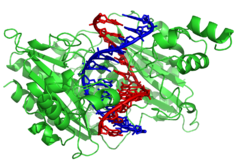
These non-specific interactions are formed through basic residues in the histones, making ionic bonds to the acidic sugar-phosphate backbone of the DNA, and are thus largely independent of the base sequence.
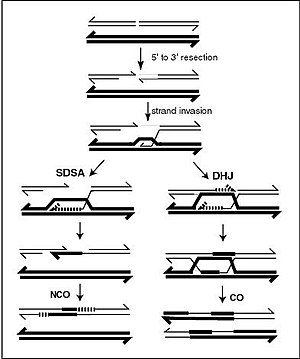
Recombination allows chromosomes to exchange genetic information and produces new combinations of genes, which increases the efficiency of natural selection and can be important in the rapid evolution of new proteins.
The formation of a Holliday junction during recombination makes it possible for genetic diversity, genes to exchange on chromosomes, and expression of wild-type viral genomes.
Pyrimidine, like polycyclic aromatic hydrocarbons (PAHs), the most carbon-rich chemical found in the universe, may have been formed in red giants or in interstellar cosmic dust and gas clouds.
[157][158] Methods have been developed to purify DNA from organisms, such as phenol-chloroform extraction, and to manipulate it in the laboratory, such as restriction digests and the polymerase chain reaction.
[165] DNA profiling was developed in 1984 by British geneticist Sir Alec Jeffreys,[166] and first used in forensic science to convict Colin Pitchfork in the 1988 Enderby murders case.
[167] The development of forensic science and the ability to now obtain genetic matching on minute samples of blood, skin, saliva, or hair has led to re-examining many cases.
Combined with the removal of the double jeopardy law in some places, this can allow cases to be reopened where prior trials have failed to produce sufficient evidence to convince a jury.
[180] Nanomechanical devices and algorithmic self-assembly have also been demonstrated,[181] and these DNA structures have been used to template the arrangement of other molecules such as gold nanoparticles and streptavidin proteins.
[182] DNA and other nucleic acids are the basis of aptamers, synthetic oligonucleotide ligands for specific target molecules used in a range of biotechnology and biomedical applications.
DNA was first isolated by the Swiss physician Friedrich Miescher who, in 1869, discovered a microscopic substance in the pus of discarded surgical bandages.
In 1933, while studying virgin sea urchin eggs, Jean Brachet suggested that DNA is found in the cell nucleus and that RNA is present exclusively in the cytoplasm.
[201] In 1943, Oswald Avery, along with co-workers Colin MacLeod and Maclyn McCarty, identified DNA as the transforming principle, supporting Griffith's suggestion (Avery–MacLeod–McCarty experiment).
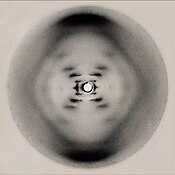
[205] In May 1952, Raymond Gosling, a graduate student working under the supervision of Rosalind Franklin, took an X-ray diffraction image, labeled as "Photo 51",[206] at high hydration levels of DNA.
[207] In February 1953, Linus Pauling and Robert Corey proposed a model for nucleic acids containing three intertwined chains, with the phosphates near the axis, and the bases on the outside.
On 28 February 1953 Crick interrupted patrons' lunchtime at The Eagle pub in Cambridge, England to announce that he and Watson had "discovered the secret of life".
[209] The 25 April 1953 issue of the journal Nature published a series of five articles giving the Watson and Crick double-helix structure DNA and evidence supporting it.
[216] In an influential presentation in 1957, Crick laid out the central dogma of molecular biology, which foretold the relationship between DNA, RNA, and proteins, and articulated the "adaptor hypothesis".
[220] In 1986, DNA analysis was first used for criminal investigative purposes when police in the UK requested Alec Jeffreys of the University of Leicester to verify or disprove a suspect's rape-murder "confession".